

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
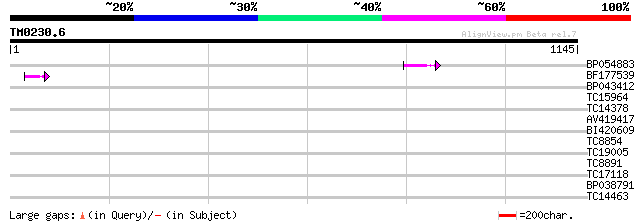
Query= TM0230.6
(1145 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BP054883 59 6e-09
BF177539 42 7e-04
BP043412 33 0.26
TC15964 similar to UP|Q8S902 (Q8S902) Syringolide-induced protei... 31 1.3
TC14378 similar to UP|O23254 (O23254) Serine hydroxymethyltransf... 31 1.3
AV419417 30 1.7
BI420609 30 2.2
TC8854 weakly similar to UP|Q9LEU4 (Q9LEU4) CCR4-associated fact... 30 2.8
TC19005 weakly similar to UP|O23617 (O23617) Trehalose-6-phospha... 29 4.8
TC8891 similar to GB|AAP68279.1|31711846|BT008840 At1g72680 {Ara... 28 6.3
TC17118 similar to UP|Q94CL1 (Q94CL1) RING finger-like protein (... 28 6.3
BP038791 28 8.2
TC14463 similar to UP|Q9SLF1 (Q9SLF1) Nodulin-like protein (At2g... 28 8.2
>BP054883
Length = 545
Score = 58.5 bits (140), Expect = 6e-09
Identities = 34/77 (44%), Positives = 46/77 (59%), Gaps = 1/77 (1%)
Frame = -3
Query: 795 RNKAYPEGSIAESFLANECLTFCSQYL-SGVETRFSRPTRNDDEFDSTLDDGSFLHPMGR 853
RNK+ PE SIAE++L ECLT CS+YL GVETR +R RN D +L HP+G
Sbjct: 543 RNKSRPEVSIAEAYLVEECLTLCSRYLHGGVETRLNRMRRNGDRSHPSLG-----HPIG- 382
Query: 854 PLGRKRQQKFQLKKRKR 870
G+K+ + L + +
Sbjct: 381 --GKKKGEVISLNYKSK 337
>BF177539
Length = 327
Score = 41.6 bits (96), Expect = 7e-04
Identities = 23/51 (45%), Positives = 26/51 (50%)
Frame = +3
Query: 30 FQNSQVSEKIWCPCTTCANCKSLSRTSVYEHLTNPRRGFLRGYKQWIFHGE 80
F+ S V I CPC C K L+R VYEHL + F R Y W HGE
Sbjct: 15 FERSSVDGAILCPCGLCNFRKWLTRDVVYEHLI--IKQFPRNYTFWFDHGE 161
>BP043412
Length = 467
Score = 33.1 bits (74), Expect = 0.26
Identities = 16/45 (35%), Positives = 26/45 (57%)
Frame = -1
Query: 700 KVLNVKFIEQLESQIAITLCQLETIFPPSFFTIMVHLVVHLAHEA 744
K++NV+ I + E ++ ++ IF P I+ HL+VHL H A
Sbjct: 350 KMMNVREISREEEEVKGRSQEMRPIFTPGMEDILHHLLVHLGHIA 216
>TC15964 similar to UP|Q8S902 (Q8S902) Syringolide-induced protein 19-1-5,
partial (27%)
Length = 401
Score = 30.8 bits (68), Expect = 1.3
Identities = 11/28 (39%), Positives = 14/28 (49%)
Frame = +3
Query: 37 EKIWCPCTTCANCKSLSRTSVYEHLTNP 64
E WC C C +C S + S+ L NP
Sbjct: 168 ETSWCHCNNCKSCNSSAPNSICHDLLNP 251
>TC14378 similar to UP|O23254 (O23254) Serine hydroxymethyltransferase
(Serine methylase) (Glycine hydroxymethyltransferase)
(SHMT) , partial (68%)
Length = 1023
Score = 30.8 bits (68), Expect = 1.3
Identities = 11/18 (61%), Positives = 13/18 (72%)
Frame = +2
Query: 448 SGWSTKGKYACPCCGFET 465
SGWS++ CPCC FET
Sbjct: 926 SGWSSQSPDWCPCCRFET 979
>AV419417
Length = 405
Score = 30.4 bits (67), Expect = 1.7
Identities = 15/46 (32%), Positives = 24/46 (51%)
Frame = -1
Query: 775 IIYI*NYFFPRFLLTLKSFVRNKAYPEGSIAESFLANECLTFCSQY 820
I Y + + +FLL + ++RNK +P + FL C+ F S Y
Sbjct: 180 IKYFRSINYDQFLLVVPYYLRNKDFPSWCVCFIFLNVMCVLFLSNY 43
>BI420609
Length = 543
Score = 30.0 bits (66), Expect = 2.2
Identities = 15/60 (25%), Positives = 26/60 (43%)
Frame = +3
Query: 604 SPNTNTMLLPRACYQMTTKEKDDFLSVLKNVKAPDECSSNISRCVQVQQRKIFGLKSYDC 663
S + N L+ R C++ + +DD L CS ++ C + + G K +DC
Sbjct: 267 SVSANCHLIERLCFESSKAGRDDSLKA-------QTCSELVNNCPHLASLSLRGFKLHDC 425
>TC8854 weakly similar to UP|Q9LEU4 (Q9LEU4) CCR4-associated factor-like
protein, partial (8%)
Length = 916
Score = 29.6 bits (65), Expect = 2.8
Identities = 14/37 (37%), Positives = 20/37 (53%), Gaps = 2/37 (5%)
Frame = +3
Query: 450 WSTKGKYACP--CCGFETSSQWLRNGRKFCYMSHRRW 484
W+TK CP CC +E ++W+R FC + R W
Sbjct: 600 WATK*NTGCPEGCC-WEEVTKWVRTVC*FCMLLKRSW 707
>TC19005 weakly similar to UP|O23617 (O23617) Trehalose-6-phosphate
synthase homolog (Trehalose-6-phosphate synthase like
protein), partial (22%)
Length = 1125
Score = 28.9 bits (63), Expect = 4.8
Identities = 8/13 (61%), Positives = 10/13 (76%)
Frame = -2
Query: 40 WCPCTTCANCKSL 52
WCPCT+C+ C L
Sbjct: 80 WCPCTSCSRCLQL 42
>TC8891 similar to GB|AAP68279.1|31711846|BT008840 At1g72680 {Arabidopsis
thaliana;} , complete
Length = 1636
Score = 28.5 bits (62), Expect = 6.3
Identities = 10/18 (55%), Positives = 14/18 (77%)
Frame = +1
Query: 428 FQLHAALMWTINDFPAYA 445
F +HAA+ WT++D AYA
Sbjct: 1525 FSIHAAIYWTVDDVIAYA 1578
>TC17118 similar to UP|Q94CL1 (Q94CL1) RING finger-like protein
(AT5g19430/F7K24_180), partial (35%)
Length = 708
Score = 28.5 bits (62), Expect = 6.3
Identities = 11/23 (47%), Positives = 12/23 (51%)
Frame = +2
Query: 215 HACPNDCILYWDKYTNDEVCPKC 237
HA CIL W Y N CP+C
Sbjct: 401 HAYCVTCILRWATYNNKVTCPQC 469
>BP038791
Length = 447
Score = 28.1 bits (61), Expect = 8.2
Identities = 18/50 (36%), Positives = 23/50 (46%), Gaps = 8/50 (16%)
Frame = -2
Query: 123 ERNPQVEENANKFYRLVKDNE--------QILYPNCKKYSKLSFMVHLYH 164
+R P VE+ K V D E Q+ P C+ KL F+VHL H
Sbjct: 149 KREPDVEDCDGKMVMTV-D*ELCKFQG*GQVAMPRCRNEDKLCFLVHLTH 3
>TC14463 similar to UP|Q9SLF1 (Q9SLF1) Nodulin-like protein
(At2g16660/T24I21.7), partial (40%)
Length = 1122
Score = 28.1 bits (61), Expect = 8.2
Identities = 12/27 (44%), Positives = 13/27 (47%)
Frame = -3
Query: 458 CPCCGFETSSQWLRNGRKFCYMSHRRW 484
C CC S+W R GR C RRW
Sbjct: 202 CSCCRRWIGSRWRRRGRAIC----RRW 134
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.336 0.147 0.497
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 24,369,169
Number of Sequences: 28460
Number of extensions: 395538
Number of successful extensions: 3338
Number of sequences better than 10.0: 26
Number of HSP's better than 10.0 without gapping: 3297
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3337
length of query: 1145
length of database: 4,897,600
effective HSP length: 100
effective length of query: 1045
effective length of database: 2,051,600
effective search space: 2143922000
effective search space used: 2143922000
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.7 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0230.6


