

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0181.1
(325 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
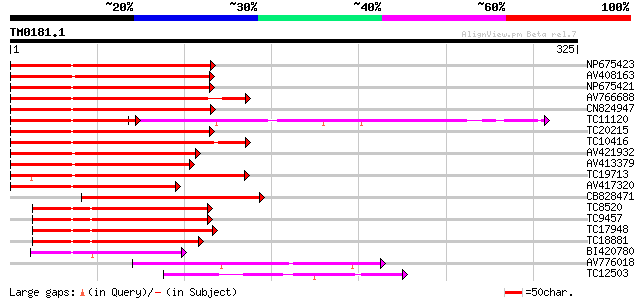
Score E
Sequences producing significant alignments: (bits) Value
NP675423 transcription factor MYB101 [Lotus japonicus] 172 5e-44
AV408163 162 5e-41
NP675421 transcription factor MYB103 [Lotus japonicus] 159 8e-40
AV766688 158 1e-39
CN824947 156 4e-39
TC11120 UP|Q84PP4 (Q84PP4) Transcription factor MYB102, complete 110 4e-39
TC20215 similar to AAS10104 (AAS10104) MYB transcription factor,... 155 7e-39
TC10416 weakly similar to UP|O04108 (O04108) MYB factor, partial... 152 6e-38
AV421932 145 7e-36
AV413379 137 2e-33
TC19713 homologue to UP|P93474 (P93474) Myb26, partial (55%) 130 2e-31
AV417320 129 5e-31
CB828471 119 7e-28
TC8520 homologue to UP|Q9FZ15 (Q9FZ15) Tuber-specific and sucros... 107 2e-24
TC9457 homologue to UP|Q9FZ15 (Q9FZ15) Tuber-specific and sucros... 107 3e-24
TC17948 similar to UP|Q9FZ13 (Q9FZ13) Tuber-specific and sucrose... 104 2e-23
TC18881 similar to UP|Q9FZ15 (Q9FZ15) Tuber-specific and sucrose... 99 7e-22
BI420780 77 4e-15
AV776018 66 9e-12
TC12503 homologue to UP|Q9ATD9 (Q9ATD9) BNLGHi233, partial (24%) 56 9e-09
>NP675423 transcription factor MYB101 [Lotus japonicus]
Length = 1047
Score = 172 bits (437), Expect = 5e-44
Identities = 77/118 (65%), Positives = 93/118 (78%)
Frame = +1
Query: 1 MGRAPCCDKANVKRGPWSPEEDAKLKAYIDHHGTGGNWIALPQKIGLKRCGKSCRLRWLN 60
MGR PCCD+ +K+GPW+PEEDA L YI HG G +W ALP+ GL RCGKSCRLRW N
Sbjct: 1 MGRTPCCDEIGLKKGPWTPEEDAILVDYIQKHGHG-SWRALPKLAGLNRCGKSCRLRWTN 177
Query: 61 YLRPNIKHGGFSEEEDNIICNLYVSIGSRWSIIAAQLPGRTDNDIKNYWNTRLKKKLL 118
YLRP+IK G F+EEE+ +I NL+ +G++WS IA LPGRTDN+IKN+WNT LKKKLL
Sbjct: 178 YLRPDIKRGKFTEEEEQLIINLHAVLGNKWSAIAGHLPGRTDNEIKNFWNTHLKKKLL 351
>AV408163
Length = 425
Score = 162 bits (411), Expect = 5e-41
Identities = 69/117 (58%), Positives = 94/117 (79%)
Frame = +2
Query: 1 MGRAPCCDKANVKRGPWSPEEDAKLKAYIDHHGTGGNWIALPQKIGLKRCGKSCRLRWLN 60
MGR+PCC+KA++ +G W+ EED +L +YI HG G W +LP+ GL RCGKSCRLRW+N
Sbjct: 77 MGRSPCCEKAHINKGAWTKEEDDRLISYIRAHGEGC-WRSLPKAAGLLRCGKSCRLRWIN 253
Query: 61 YLRPNIKHGGFSEEEDNIICNLYVSIGSRWSIIAAQLPGRTDNDIKNYWNTRLKKKL 117
YLRP++K G F+EEED +I L+ +G++WS+IA +LPGRTDN+IKNYWNT +++KL
Sbjct: 254 YLRPDLKRGNFTEEEDELIIKLHSLLGNKWSLIAGRLPGRTDNEIKNYWNTHIRRKL 424
>NP675421 transcription factor MYB103 [Lotus japonicus]
Length = 924
Score = 159 bits (401), Expect = 8e-40
Identities = 73/117 (62%), Positives = 89/117 (75%)
Frame = +1
Query: 1 MGRAPCCDKANVKRGPWSPEEDAKLKAYIDHHGTGGNWIALPQKIGLKRCGKSCRLRWLN 60
M R P D++ +K+GPWS EED L YI HG G +W ALPQ GL RCGKSCRLRW N
Sbjct: 1 MMRTPSIDESGMKKGPWSQEEDKVLVDYIQKHGHG-SWRALPQLAGLNRCGKSCRLRWTN 177
Query: 61 YLRPNIKHGGFSEEEDNIICNLYVSIGSRWSIIAAQLPGRTDNDIKNYWNTRLKKKL 117
YLRP+IK G FS+EE+ +I NL+ S+G++W+ IA+ LPGRTDN+IKN WNT LKKKL
Sbjct: 178 YLRPDIKRGKFSDEEEQLIINLHASLGNKWATIASHLPGRTDNEIKNLWNTHLKKKL 348
>AV766688
Length = 608
Score = 158 bits (399), Expect = 1e-39
Identities = 74/138 (53%), Positives = 99/138 (71%)
Frame = +2
Query: 1 MGRAPCCDKANVKRGPWSPEEDAKLKAYIDHHGTGGNWIALPQKIGLKRCGKSCRLRWLN 60
M R P CDK+ +++G W+ EED KL AY+ +G NW LP+ GL RCGKSCRLRWLN
Sbjct: 77 MARTPSCDKSGLRKGTWTAEEDRKLIAYVTRYGCW-NWRQLPKFAGLARCGKSCRLRWLN 253
Query: 61 YLRPNIKHGGFSEEEDNIICNLYVSIGSRWSIIAAQLPGRTDNDIKNYWNTRLKKKLLGK 120
YLRP+IK G F++EE+ II ++ ++G+RWSIIAA+LPGRTDN++KN+W+T LK
Sbjct: 254 YLRPDIKRGHFTQEEEEIIIRMHKNLGNRWSIIAAELPGRTDNEVKNHWHTSLK------ 415
Query: 121 QRKEQQAQARRVSTMKQE 138
R+ QQ + +VS K E
Sbjct: 416 -RRVQQNEEAKVSNSKNE 466
>CN824947
Length = 738
Score = 156 bits (395), Expect = 4e-39
Identities = 68/118 (57%), Positives = 90/118 (75%)
Frame = +1
Query: 1 MGRAPCCDKANVKRGPWSPEEDAKLKAYIDHHGTGGNWIALPQKIGLKRCGKSCRLRWLN 60
MGRAPCC+K +KRG W+ EED L YI G G +W +LP+ GL RCGKSCRLRW+N
Sbjct: 151 MGRAPCCEKVGLKRGRWTAEEDELLTKYIQASGEG-SWRSLPKNAGLLRCGKSCRLRWIN 327
Query: 61 YLRPNIKHGGFSEEEDNIICNLYVSIGSRWSIIAAQLPGRTDNDIKNYWNTRLKKKLL 118
YLR ++K G S+EE+++I L+ S G+RWS+IA+ +PGRTDN+IKNYWN+ L +KL+
Sbjct: 328 YLRGDLKRGNISDEEESLIVKLHASFGNRWSLIASHMPGRTDNEIKNYWNSHLSRKLV 501
>TC11120 UP|Q84PP4 (Q84PP4) Transcription factor MYB102, complete
Length = 1281
Score = 110 bits (275), Expect(2) = 4e-39
Identities = 49/75 (65%), Positives = 58/75 (77%)
Frame = +1
Query: 1 MGRAPCCDKANVKRGPWSPEEDAKLKAYIDHHGTGGNWIALPQKIGLKRCGKSCRLRWLN 60
MGR+PCCD+ +K+GPW+PEED KL +I HG G +W ALP+ GL RCGKSCRLRW N
Sbjct: 67 MGRSPCCDENGLKKGPWTPEEDQKLVEHIQKHGHG-SWRALPKLAGLNRCGKSCRLRWTN 243
Query: 61 YLRPNIKHGGFSEEE 75
YLRP+IK G FS EE
Sbjct: 244 YLRPDIKRGKFSPEE 288
Score = 67.4 bits (163), Expect(2) = 4e-39
Identities = 67/257 (26%), Positives = 109/257 (42%), Gaps = 16/257 (6%)
Frame = +2
Query: 69 GGFSEEEDNIICNLYVSIGSRWSIIAAQLPGRTDNDIKNYWNTRLKKKL--LGKQRKEQQ 126
G F ++++ I +L+ +G++WS IA LPGRTDN+IKN+WNT LKKKL +G Q
Sbjct: 269 GNFLQKKEQTILHLHSILGNKWSTIATHLPGRTDNEIKNFWNTHLKKKLIQMGFDPMTHQ 448
Query: 127 AQARRVSTMKQEIKRETDQNLMVASGVNTTHAAFYWPAEYSIPMPVANASIQ-------- 178
+ VST+ + LM + H +F A + N +Q
Sbjct: 449 PRTDLVSTLPYLLALANMTQLM-----DHNHQSFDEQAVQLAKLQYLNYLLQSSNNPLCM 613
Query: 179 --DYDFHNQTSLKSL-LINKQGGVR--FSYDHQQPYSNAITATANSQDPCDIYPASTMNM 233
YD + T++++ L+N V+ D Q + + + + P +
Sbjct: 614 TNSYDQNALTNMEAFSLLNSISNVKENLGVDFSQMDTQTSLFSHDGNNMASSQPLHHPSN 793
Query: 234 INPVAMVSNANIGGTTCQQSN-VFQGFENFTSDFSELVCVNPQTIDGFYGMESLEMSNVS 292
+ P + + +C +N QG NF ++ + G + N+
Sbjct: 794 MLPHFLDPQVSFSSQSCLNNNEQGQGTNNF--------AISQRDHTG----DESSWINIP 937
Query: 293 STNTPSAESTSWGDMSS 309
S N P+A TS GD SS
Sbjct: 938 SINPPNAIGTS-GDASS 985
>TC20215 similar to AAS10104 (AAS10104) MYB transcription factor, partial
(35%)
Length = 464
Score = 155 bits (393), Expect = 7e-39
Identities = 67/117 (57%), Positives = 89/117 (75%)
Frame = +2
Query: 1 MGRAPCCDKANVKRGPWSPEEDAKLKAYIDHHGTGGNWIALPQKIGLKRCGKSCRLRWLN 60
MGRAPCC+K +KRG WS EED L +I +G G +W +LP+ GL RCGKSCRLRW+N
Sbjct: 29 MGRAPCCEKVGLKRGRWSEEEDKILTDFIKQNGEG-SWKSLPKNAGLLRCGKSCRLRWIN 205
Query: 61 YLRPNIKHGGFSEEEDNIICNLYVSIGSRWSIIAAQLPGRTDNDIKNYWNTRLKKKL 117
YLR ++K G + EE+ II L+ ++G+RWS+IA LPGRTDN+IKNYWN+ L++K+
Sbjct: 206 YLREDVKRGNITSEEEEIIVKLHAALGNRWSVIAGHLPGRTDNEIKNYWNSHLRRKI 376
>TC10416 weakly similar to UP|O04108 (O04108) MYB factor, partial (41%)
Length = 1046
Score = 152 bits (385), Expect = 6e-38
Identities = 70/138 (50%), Positives = 97/138 (69%)
Frame = +2
Query: 1 MGRAPCCDKANVKRGPWSPEEDAKLKAYIDHHGTGGNWIALPQKIGLKRCGKSCRLRWLN 60
M R P CDK+ +++G W+ EED KL AY+ +G NW LP+ GL RCGKSCRLRWLN
Sbjct: 92 MARTPSCDKSGMRKGTWTAEEDKKLIAYVTRYGCW-NWRQLPKFAGLARCGKSCRLRWLN 268
Query: 61 YLRPNIKHGGFSEEEDNIICNLYVSIGSRWSIIAAQLPGRTDNDIKNYWNTRLKKKLLGK 120
YLRPNIK G F++EE+ II ++ +G+RWS IAA+LPGRTD+++KN+W+T LK+ +
Sbjct: 269 YLRPNIKRGNFTQEEEEIIIRMHKKLGNRWSTIAAELPGRTDSEVKNHWHTTLKRTV--H 442
Query: 121 QRKEQQAQARRVSTMKQE 138
++ + +VS K E
Sbjct: 443 EQNTVTNEETKVSNSKNE 496
>AV421932
Length = 402
Score = 145 bits (367), Expect = 7e-36
Identities = 65/109 (59%), Positives = 83/109 (75%)
Frame = +3
Query: 1 MGRAPCCDKANVKRGPWSPEEDAKLKAYIDHHGTGGNWIALPQKIGLKRCGKSCRLRWLN 60
MG CC++ VKRG WSPEED KL YI HG G W +P+K GL+RCGKSCRLRW+N
Sbjct: 78 MGHHSCCNQQKVKRGLWSPEEDEKLIRYITTHGYGC-WSEVPEKAGLQRCGKSCRLRWIN 254
Query: 61 YLRPNIKHGGFSEEEDNIICNLYVSIGSRWSIIAAQLPGRTDNDIKNYW 109
YLRP+I+ G F+ EE+ +I +L+ +G+RW+ IA+ LPGRTDN+IKNYW
Sbjct: 255 YLRPDIRRGRFTPEEEKLIISLHGVVGNRWAHIASHLPGRTDNEIKNYW 401
>AV413379
Length = 416
Score = 137 bits (345), Expect = 2e-33
Identities = 60/106 (56%), Positives = 81/106 (75%)
Frame = +3
Query: 1 MGRAPCCDKANVKRGPWSPEEDAKLKAYIDHHGTGGNWIALPQKIGLKRCGKSCRLRWLN 60
MGR+PCC+KA+ +G W+ EED +L AYI HG G W +LP+ GL RCGKSCRLRW+N
Sbjct: 102 MGRSPCCEKAHTNKGAWTKEEDDRLIAYIRAHGEGC-WRSLPKSAGLLRCGKSCRLRWIN 278
Query: 61 YLRPNIKHGGFSEEEDNIICNLYVSIGSRWSIIAAQLPGRTDNDIK 106
YLRP++K G F+ +ED +I L+ +G++WS+IA +L GRTDN+IK
Sbjct: 279 YLRPDLKRGNFTPQEDELIIKLHSLLGNKWSLIAGRLAGRTDNEIK 416
>TC19713 homologue to UP|P93474 (P93474) Myb26, partial (55%)
Length = 532
Score = 130 bits (328), Expect = 2e-31
Identities = 64/139 (46%), Positives = 88/139 (63%), Gaps = 2/139 (1%)
Frame = +3
Query: 1 MGRAPCCDKAN--VKRGPWSPEEDAKLKAYIDHHGTGGNWIALPQKIGLKRCGKSCRLRW 58
M + PC + V++GPW+ EED L YI +HG G W L + GLKR GKSCRLRW
Sbjct: 66 MDKKPCNSSHDPEVRKGPWTIEEDLILINYIANHGEGV-WNTLAKSAGLKRTGKSCRLRW 242
Query: 59 LNYLRPNIKHGGFSEEEDNIICNLYVSIGSRWSIIAAQLPGRTDNDIKNYWNTRLKKKLL 118
LNYLRP+++ G + EE +I L+ G+RWS IA LPGRTDN+IKN+W TR++K +
Sbjct: 243 LNYLRPDVRRGNITPEEQLLIMELHAKWGNRWSKIAKHLPGRTDNEIKNFWRTRIQKHIK 422
Query: 119 GKQRKEQQAQARRVSTMKQ 137
+ +QQ + ++ Q
Sbjct: 423 QAESLQQQQSSSEINDHHQ 479
>AV417320
Length = 369
Score = 129 bits (325), Expect = 5e-31
Identities = 58/98 (59%), Positives = 74/98 (75%)
Frame = +3
Query: 1 MGRAPCCDKANVKRGPWSPEEDAKLKAYIDHHGTGGNWIALPQKIGLKRCGKSCRLRWLN 60
M RAPCC+K +K+GPW+ EED L +YI HG G NW ALP+ GL RCGKSCRLRW+N
Sbjct: 78 MVRAPCCEKIGLKKGPWTSEEDQILISYIQKHGHG-NWRALPKHAGLLRCGKSCRLRWIN 254
Query: 61 YLRPNIKHGGFSEEEDNIICNLYVSIGSRWSIIAAQLP 98
YLRP+IK G F+ EE+ +I ++ +G+RWS IAA+LP
Sbjct: 255 YLRPDIKRGNFTAEEEELIIKMHELLGNRWSAIAAKLP 368
>CB828471
Length = 525
Score = 119 bits (298), Expect = 7e-28
Identities = 55/105 (52%), Positives = 76/105 (72%)
Frame = +1
Query: 42 PQKIGLKRCGKSCRLRWLNYLRPNIKHGGFSEEEDNIICNLYVSIGSRWSIIAAQLPGRT 101
P++ GL RCGKSCRLRW NYLRP+IK G FS EE++ I NL+ +G+RWS IAA+LPGRT
Sbjct: 1 PKQAGLLRCGKSCRLRWANYLRPDIKRGNFSREEEDEIINLHELLGNRWSAIAARLPGRT 180
Query: 102 DNDIKNYWNTRLKKKLLGKQRKEQQAQARRVSTMKQEIKRETDQN 146
DN+IKN W+T LKK+L + +++Q S ++ K+E ++
Sbjct: 181 DNEIKNVWHTHLKKRLQPQTKQKQTNLDLDASKSNKDAKKEHQED 315
>TC8520 homologue to UP|Q9FZ15 (Q9FZ15) Tuber-specific and
sucrose-responsive element binding factor, partial (36%)
Length = 1778
Score = 107 bits (268), Expect = 2e-24
Identities = 50/103 (48%), Positives = 67/103 (64%)
Frame = +1
Query: 14 RGPWSPEEDAKLKAYIDHHGTGGNWIALPQKIGLKRCGKSCRLRWLNYLRPNIKHGGFSE 73
+GPWSPEED L+ ++ HG NW + + I R GKSCRLRW N L P ++H F+
Sbjct: 184 KGPWSPEEDEALQKLVERHGPR-NWSLISKSIP-GRSGKSCRLRWCNQLSPQVEHRAFTP 357
Query: 74 EEDNIICNLYVSIGSRWSIIAAQLPGRTDNDIKNYWNTRLKKK 116
EED+ I + G++W+ IA L GRTDN IKN+WN+ LK+K
Sbjct: 358 EEDDTIIRAHARFGNKWATIARLLSGRTDNAIKNHWNSTLKRK 486
Score = 30.8 bits (68), Expect = 0.33
Identities = 16/50 (32%), Positives = 26/50 (52%), Gaps = 1/50 (2%)
Frame = +1
Query: 69 GGFSEEEDNIICNLYVSIGSR-WSIIAAQLPGRTDNDIKNYWNTRLKKKL 117
G +S EED + L G R WS+I+ +PGR+ + W +L ++
Sbjct: 187 GPWSPEEDEALQKLVERHGPRNWSLISKSIPGRSGKSCRLRWCNQLSPQV 336
>TC9457 homologue to UP|Q9FZ15 (Q9FZ15) Tuber-specific and
sucrose-responsive element binding factor, partial (43%)
Length = 860
Score = 107 bits (267), Expect = 3e-24
Identities = 50/103 (48%), Positives = 67/103 (64%)
Frame = +3
Query: 14 RGPWSPEEDAKLKAYIDHHGTGGNWIALPQKIGLKRCGKSCRLRWLNYLRPNIKHGGFSE 73
+GPWSPEED L+ ++ HG NW + + I R GKSCRLRW N L P ++H F+
Sbjct: 78 KGPWSPEEDEALQRLVEKHGPR-NWSLISKSIP-GRSGKSCRLRWCNQLSPQVEHRAFTA 251
Query: 74 EEDNIICNLYVSIGSRWSIIAAQLPGRTDNDIKNYWNTRLKKK 116
EED+ I + G++W+ IA L GRTDN IKN+WN+ LK+K
Sbjct: 252 EEDDTIIRAHARFGNKWATIARLLSGRTDNAIKNHWNSTLKRK 380
Score = 31.2 bits (69), Expect = 0.25
Identities = 16/50 (32%), Positives = 26/50 (52%), Gaps = 1/50 (2%)
Frame = +3
Query: 69 GGFSEEEDNIICNLYVSIGSR-WSIIAAQLPGRTDNDIKNYWNTRLKKKL 117
G +S EED + L G R WS+I+ +PGR+ + W +L ++
Sbjct: 81 GPWSPEEDEALQRLVEKHGPRNWSLISKSIPGRSGKSCRLRWCNQLSPQV 230
>TC17948 similar to UP|Q9FZ13 (Q9FZ13) Tuber-specific and sucrose-responsive
element binding factor, partial (50%)
Length = 487
Score = 104 bits (260), Expect = 2e-23
Identities = 50/106 (47%), Positives = 67/106 (63%)
Frame = +2
Query: 14 RGPWSPEEDAKLKAYIDHHGTGGNWIALPQKIGLKRCGKSCRLRWLNYLRPNIKHGGFSE 73
+GPWS EED L ++ HG NW + + I R GKSCRLRW N L P ++H FS
Sbjct: 170 KGPWSAEEDRILTRLVERHGAR-NWSLISRYIK-GRSGKSCRLRWCNQLSPTVEHRPFSV 343
Query: 74 EEDNIICNLYVSIGSRWSIIAAQLPGRTDNDIKNYWNTRLKKKLLG 119
+ED I + G+RW+ IA LPGRTDN +KN+WN+ LK+++ G
Sbjct: 344 QEDETIIAAHARFGNRWAAIARLLPGRTDNAVKNHWNSTLKRRVPG 481
>TC18881 similar to UP|Q9FZ15 (Q9FZ15) Tuber-specific and sucrose-responsive
element binding factor, partial (28%)
Length = 346
Score = 99.4 bits (246), Expect = 7e-22
Identities = 47/98 (47%), Positives = 62/98 (62%)
Frame = +1
Query: 14 RGPWSPEEDAKLKAYIDHHGTGGNWIALPQKIGLKRCGKSCRLRWLNYLRPNIKHGGFSE 73
+GPWSPEED L + HG NW A+ + I R GKSCRLRW N L P ++H F+
Sbjct: 55 KGPWSPEEDEALTRLVQVHGPK-NWTAISKSIP-GRSGKSCRLRWCNQLSPEVEHRPFTP 228
Query: 74 EEDNIICNLYVSIGSRWSIIAAQLPGRTDNDIKNYWNT 111
EED+ I + G++W+ IA L GRTDN +KN+WN+
Sbjct: 229 EEDSTIIRAHARCGNKWATIARILNGRTDNAVKNHWNS 342
>BI420780
Length = 475
Score = 77.0 bits (188), Expect = 4e-15
Identities = 39/91 (42%), Positives = 55/91 (59%), Gaps = 2/91 (2%)
Frame = +1
Query: 13 KRGPWSPEEDAKLKAYIDHHGTGGNWIALPQKIG--LKRCGKSCRLRWLNYLRPNIKHGG 70
+R WS EEDA L AY+ +G W + Q++ L R KSC RW NYL+P IK G
Sbjct: 199 ERQRWSSEEDALLHAYVQQYGPR-EWNLVSQRMNTPLNRDTKSCLERWKNYLKPGIKKGS 375
Query: 71 FSEEEDNIICNLYVSIGSRWSIIAAQLPGRT 101
++EE ++ L + G++W IAA++PGRT
Sbjct: 376 LTKEEQRLVILLQANYGNKWKKIAAEVPGRT 468
>AV776018
Length = 476
Score = 65.9 bits (159), Expect = 9e-12
Identities = 46/156 (29%), Positives = 76/156 (48%), Gaps = 11/156 (7%)
Frame = +1
Query: 71 FSEEEDNIICNLYVSIGSRWSIIAAQLPGRTDNDIKNYWNTRLKKKLLGK------QRKE 124
F +EEDN+I L+ +G+RWS A Q PGRTDN+IKN WN+ LKKKL + +
Sbjct: 13 FHKEEDNLIIELHAVLGNRWSQDATQWPGRTDNEIKNLWNSCLKKKLRQRGIDPVTHKPL 192
Query: 125 QQAQARRVSTMKQEIKRETDQNLMVASGVNTTHAAFYWPAEYSIPMPVANASIQDYDFHN 184
+ + + K + K + NL+ + + A++ S P A + + +
Sbjct: 193 SEVENGKEDKNKSQEKVSNELNLLKSESFKSDAASY--EQNSSSLAPKAYTTEMEGSSSS 366
Query: 185 QTSLKSLLINK-----QGGVRFSYDHQQPYSNAITA 215
S K LL+++ Q G+ Y Q +++ +A
Sbjct: 367 DCSSKDLLLDRFMSQTQNGMGNYYPLQMSFASTDSA 474
>TC12503 homologue to UP|Q9ATD9 (Q9ATD9) BNLGHi233, partial (24%)
Length = 957
Score = 55.8 bits (133), Expect = 9e-09
Identities = 42/143 (29%), Positives = 66/143 (45%), Gaps = 3/143 (2%)
Frame = +2
Query: 89 RWSIIAAQLPGRTDNDIKNYWNTRLKKKLLGKQRKEQQAQARRVSTMKQEIKRETDQNLM 148
RWS+IA ++PGRTDN+IKN+WNT L KKL+ + + + + L
Sbjct: 152 RWSLIAGRIPGRTDNEIKNFWNTHLSKKLISQ-------------GIDPRTHKPLNPELS 292
Query: 149 VASGVNTTHAAFYWPAEYSIPMPVA--NASIQDYDFHNQTSLKSLLINKQGGVRFSYDHQ 206
+ S T P S P+PV + ++ + H+ ++ S+ G Y +Q
Sbjct: 293 IPSSSTTPP-----PPPPSKPLPVITNHQTLHETTTHHHLNIASVNQECDEG---QYQYQ 448
Query: 207 QPYSNAITATANSQD-PCDIYPA 228
QP + A A + D P D P+
Sbjct: 449 QPQAVAAAAYMVANDIPDDDVPS 517
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.316 0.130 0.399
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,038,483
Number of Sequences: 28460
Number of extensions: 83468
Number of successful extensions: 425
Number of sequences better than 10.0: 50
Number of HSP's better than 10.0 without gapping: 403
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 408
length of query: 325
length of database: 4,897,600
effective HSP length: 90
effective length of query: 235
effective length of database: 2,336,200
effective search space: 549007000
effective search space used: 549007000
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 55 (25.8 bits)
Lotus: description of TM0181.1


