

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0129.4
(1259 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
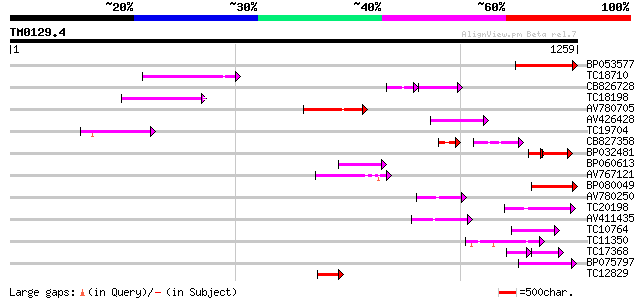
Score E
Sequences producing significant alignments: (bits) Value
BP053577 152 4e-37
TC18710 149 3e-36
CB826728 97 5e-31
TC18198 weakly similar to UP|Q9XGZ5 (Q9XGZ5) T1N24.20 protein, p... 119 4e-27
AV780705 117 1e-26
AV426428 106 2e-23
TC19704 weakly similar to UP|Q9SZ87 (Q9SZ87) RNA-directed DNA po... 104 8e-23
CB827358 74 3e-21
BP032481 69 9e-19
BP060613 89 6e-18
AV767121 88 7e-18
BP080049 87 2e-17
AV780250 86 3e-17
TC20198 similar to UP|Q9ZPU0 (Q9ZPU0) At2g13980 protein, partial... 86 3e-17
AV411435 84 2e-16
TC10764 72 7e-13
TC11350 similar to UP|Q9LJP7 (Q9LJP7) Genomic DNA, chromosome 3,... 72 7e-13
TC17368 51 7e-13
BP075797 70 2e-12
TC12829 70 2e-12
>BP053577
Length = 463
Score = 152 bits (383), Expect = 4e-37
Identities = 74/137 (54%), Positives = 94/137 (68%)
Frame = +3
Query: 1123 GEVGFGLIVRNMLGEVLASAAQYLLHAASAILGEALVFRWSMQLTIQMEFRRVLFETDCL 1182
G G G++ RN GEV+ASA + +S +L EAL RW+MQL + FRR+ ETDCL
Sbjct: 6 GIAGLGMVARNCDGEVMASACSSPVSISSPLLAEALGLRWTMQLATDLGFRRITLETDCL 185
Query: 1183 QLFQLWKKPPDGRSYLSSIVGDCFMLSRFFDYVDLSFVRRRGNSVADFLAGTASKYTDMV 1242
QLF W+ +GRSYLSSI+ DC +L FDYV +SFVRR GN+VADFLA + Y +MV
Sbjct: 186 QLFNTWRSQEEGRSYLSSIISDCRILISAFDYVSVSFVRRTGNTVADFLARNSDTYANMV 365
Query: 1243 WLEEVPREVITLIHKDV 1259
W+EEVP V++LI DV
Sbjct: 366 WVEEVPLTVVSLIDADV 416
>TC18710
Length = 843
Score = 149 bits (376), Expect = 3e-36
Identities = 87/217 (40%), Positives = 122/217 (56%), Gaps = 1/217 (0%)
Frame = +2
Query: 296 YFGGLFTSSNPEGIEETTNLVAGRVSETHLTVLGEPFTREEVEEALFQMHPTKAPGLDGF 355
+F L+TS+ P E L + + L ++++ F + ++ G+DG
Sbjct: 23 FFPELYTSTPPNSSESA*MLFLP*SLKI*IKSLNLR*LQKKLRLLFFLLETSQPRGMDGM 202
Query: 356 PTLFYQKYWDIIGDDVSAFCLQVLQGVIPPGMINQTLLVLIPKIMKPEHATQFRPISLCN 415
LFYQK D++ D V+ L L+ P IN+TL+ LIPK+ PE QFRPIS C
Sbjct: 203 NGLFYQKNGDVVKDSVT*AILAFLERAEIPNEINETLVTLIPKVPHPESINQFRPISGCT 382
Query: 416 VLFKIITKTIANRLKIILPDIICQTQSAFVPGRLIIDNALVVYECFHFMKK-RISGRNGM 474
L+K+I+K RLK +PD+I QS F+ GR I DN L+V E FH + + GRN
Sbjct: 383 FLYKVISKIFVARLKDAMPDLISPMQSGFIQGRQIQDNLLIVQEAFHAINRPGALGRNHS 562
Query: 475 MTLKLDMSKAYDRVEWSFLQGVLQKMGFPPSWVSLIM 511
+ +KLDM+KAYDRVEW FL+ L GF +WV +IM
Sbjct: 563 I-IKLDMNKAYDRVEWKFLESSLLAFGFSTNWVKMIM 670
>CB826728
Length = 509
Score = 96.7 bits (239), Expect(2) = 5e-31
Identities = 44/98 (44%), Positives = 59/98 (59%)
Frame = +1
Query: 908 WPETVDGNYCSKSGYIFVRKNFLGACSSTYSQPSLPAPLWKKFWRTSAQPRCKEVAWRVV 967
WP + DG Y SKSGY F+R ST ++P+ +WK W S+ PRCKEV WR
Sbjct: 214 WPGSSDGWYSSKSGYEFIRLENKSLLPSTSPAAAVPSLIWKTVWSASSLPRCKEVMWRAC 393
Query: 968 SSLLPIRAALHRRGMDVDPLCPVCANEEETVFHLFVSC 1005
+ LP+R+AL RRG+DVDP P E+E H+ ++C
Sbjct: 394 AGYLPVRSALKRRGLDVDPSSPWSGLEDEAEVHVLLTC 507
Score = 56.2 bits (134), Expect(2) = 5e-31
Identities = 27/71 (38%), Positives = 39/71 (54%)
Frame = +3
Query: 836 DILHDQWLPQGVPVIGSQDLMAEFGVSKVSHLIDHAAKSWKYALVDFIFHPATVSHILKI 895
DILHD WLP P++ +DL + + KV+ LI++ W LV +F P+T + L +
Sbjct: 3 DILHDAWLPNSAPLVYCEDLFVDLQLKKVADLIENG--RWNENLVSMVFSPSTTAVFLAV 176
Query: 896 HLPLNGSVDTL 906
LPL D L
Sbjct: 177 SLPLQSYEDCL 209
>TC18198 weakly similar to UP|Q9XGZ5 (Q9XGZ5) T1N24.20 protein, partial
(14%)
Length = 592
Score = 119 bits (297), Expect = 4e-27
Identities = 72/189 (38%), Positives = 105/189 (55%)
Frame = -3
Query: 249 LKHGDKNTRFFHQKANQRRKRNLIEVLKDDNGREVEDDADIARVLGDYFGGLFTSSNPEG 308
LK GD NTRFFH QRR N I +KD +G VE + +YF +++SS +
Sbjct: 578 LK*GDHNTRFFHASTIQRRDFNRILKIKDAHGIWVEGQKKVNEAAVNYFSDIYSSSPLQY 399
Query: 309 IEETTNLVAGRVSETHLTVLGEPFTREEVEEALFQMHPTKAPGLDGFPTLFYQKYWDIIG 368
+ E + V++ L + +E++EA+F M KAPG+DG LFYQ+ +I+
Sbjct: 398 LRECLAPIPKLVTDRLNHKLCAQVSTDEIKEAVFSMGDLKAPGMDGINGLFYQQNREIVK 219
Query: 369 DDVSAFCLQVLQGVIPPGMINQTLLVLIPKIMKPEHATQFRPISLCNVLFKIITKTIANR 428
+DV+ L P +N+TL+ LIPKI E QFRPIS C+ ++K+I+K I R
Sbjct: 218 NDVNNAILDFFDHGELPVELNETLVTLIPKIPHAEAIQQFRPISCCSFIYKVISKIIVAR 39
Query: 429 LKIILPDII 437
LK PD++
Sbjct: 38 LK---PDMV 21
>AV780705
Length = 524
Score = 117 bits (293), Expect = 1e-26
Identities = 60/145 (41%), Positives = 93/145 (63%), Gaps = 4/145 (2%)
Frame = -2
Query: 653 YLGLPTMIGKFKTRIFNFVKDRVWKKLKGWKESTLSRAGREVLIKSVVQAIPSYVMSCFI 712
YLGLP + G+ K+ +++++V +KL+GWKE+ L++AG+EVLIK+++QAIPSY M+
Sbjct: 484 YLGLPAIWGRNKSHSLVWIEEKVKEKLEGWKETLLNQAGKEVLIKAIIQAIPSYAMTIVH 305
Query: 713 LPDSLCADIERMVSRFYW----GGDVDKRGLQWASWKKLTRSKFDGGLGFRDFKSFNIAL 768
P + C + +V+ F+W G V+ G A K SK GG+GFRD ++ N+A
Sbjct: 304 FPKTFCNSLYALVADFWWKSQGEGAVEYTG---AVGTK*PLSKEKGGVGFRDLRTQNLAF 134
Query: 769 VAKNWWRIYTQPETLLAQVFKGVYF 793
+A+ WR+ T PE L +V K +YF
Sbjct: 133 LARQAWRVLTNPEALWVRVMKSLYF 59
>AV426428
Length = 413
Score = 106 bits (265), Expect = 2e-23
Identities = 49/130 (37%), Positives = 72/130 (54%)
Frame = +3
Query: 934 SSTYSQPSLPAPLWKKFWRTSAQPRCKEVAWRVVSSLLPIRAALHRRGMDVDPLCPVCAN 993
+S+ PSLP+ +K W+ A PRC E WR +LP++A+LH RG+ DP+CP C
Sbjct: 6 ASSSQVPSLPSVD*RKLWKAEALPRCCETGWRACLGVLPVKASLHARGLAEDPICPRCLA 185
Query: 994 EEETVFHLFVSCPVAQNFWFGSPLSLRVHGFSSMEDFLADFFRAADDDALALWQAGVYAL 1053
+T H + CP WF S L R++ M +F+ADF + AD D + +YA+
Sbjct: 186 APKTADHAVLFCP*VHPIWFASSLGFRLNQECKMHEFVADFMQVAD*DTCGEFLTILYAI 365
Query: 1054 WEMRNRVVFR 1063
W +N + FR
Sbjct: 366 WTAQNELCFR 395
>TC19704 weakly similar to UP|Q9SZ87 (Q9SZ87) RNA-directed DNA
polymerase-like protein, partial (3%)
Length = 530
Score = 104 bits (260), Expect = 8e-23
Identities = 60/174 (34%), Positives = 87/174 (49%), Gaps = 7/174 (4%)
Frame = -3
Query: 157 ECKEVIAETWGRSLTEVPDRLSDVSG-------LLGRWGKEQFGDLPRKISDGQALLQEL 209
ECKE I + W + S++SG L W + F ++I++ + L+ L
Sbjct: 528 ECKETIKKAWDNTSETASSHCSNLSGKTTNTKLALTEWHHQNFSRADKEIAELKNQLEHL 349
Query: 210 QRKDQTKEVLMAINVAERDLDSLLEQEEVWWKQRSRASWLKHGDKNTRFFHQKANQRRKR 269
Q + E I + L L QEE++W QRSR WLK GDKNT+FFH QRR+R
Sbjct: 348 QNYSNSGEDWQEIKQRKMRLKDLWRQEELFWSQRSRIKWLKSGDKNTKFFHASTVQRRER 169
Query: 270 NLIEVLKDDNGREVEDDADIARVLGDYFGGLFTSSNPEGIEETTNLVAGRVSET 323
N IE +KD G V I R + D++ ++ S+N + I E N V + E+
Sbjct: 168 NRIERIKDMQGTWVRSQLAINRDVPDFYPDIYKSTNRQNITECLNAVPRVIYES 7
>CB827358
Length = 453
Score = 73.9 bits (180), Expect(2) = 3e-21
Identities = 43/113 (38%), Positives = 62/113 (54%), Gaps = 1/113 (0%)
Frame = +2
Query: 1030 FLADFFRAADDDALALWQAGVYALWEMRNRVVFRDGEIPVPVAAMIQRCSMLAAAPVSEM 1089
FL F AD D +A Q ALWE N+++F+ E+P A IQR + ++ VS
Sbjct: 125 FLLQFLVEADSDVVASCQMASCALWEAHNKLIFQ--EVPFRCEAAIQRATSMSQVDVS-- 292
Query: 1090 TVVPRP-APLLQATWARPPYGIYKLNFDAAVATTGEVGFGLIVRNMLGEVLAS 1141
TV P P+ A W RPP G++K+NFD + + G G++ RN G +LA+
Sbjct: 293 TVDPSANGPVHDAIWRRPPRGVFKVNFDGSWKSDSVSGIGMVARNHDGLILAA 451
Score = 46.2 bits (108), Expect(2) = 3e-21
Identities = 22/48 (45%), Positives = 31/48 (63%)
Frame = +3
Query: 953 TSAQPRCKEVAWRVVSSLLPIRAALHRRGMDVDPLCPVCANEEETVFH 1000
T A PRCK++AW V R +L R+G++ DP CP+C NE T+F+
Sbjct: 3 TKALPRCKDLAWCVCC-----R*SLWRKGVNSDPSCPLCGNEF*TIFY 131
>BP032481
Length = 447
Score = 69.3 bits (168), Expect(2) = 9e-19
Identities = 36/65 (55%), Positives = 42/65 (64%)
Frame = -1
Query: 1184 LFQLWKKPPDGRSYLSSIVGDCFMLSRFFDYVDLSFVRRRGNSVADFLAGTASKYTDMVW 1243
LF+ WK SYLSSI+ DC +L FD V LSFVRR GN+VA F A A Y ++VW
Sbjct: 258 LFKSWKAKEVE*SYLSSILQDCRLLFTAFDVVSLSFVRRTGNTVAGFFARNAETYANLVW 79
Query: 1244 LEEVP 1248
EEVP
Sbjct: 78 PEEVP 64
Score = 42.4 bits (98), Expect(2) = 9e-19
Identities = 19/37 (51%), Positives = 23/37 (61%)
Frame = -2
Query: 1153 ILGEALVFRWSMQLTIQMEFRRVLFETDCLQLFQLWK 1189
+L EA+ RW MQL I + FRR+ ETDCL L K
Sbjct: 350 LLAEAMSMRWIMQLAIDLGFRRICLETDCLTFSSLGK 240
>BP060613
Length = 378
Score = 88.6 bits (218), Expect = 6e-18
Identities = 41/106 (38%), Positives = 57/106 (53%)
Frame = +1
Query: 730 WGGDVDKRGLQWASWKKLTRSKFDGGLGFRDFKSFNIALVAKNWWRIYTQPETLLAQVFK 789
W RG+ W W LT K +GGLGF+DF+ N AL+AK WRI P+ L Q+ K
Sbjct: 46 WWSSKGTRGVHWRKWDLLTELKDEGGLGFKDFEIQNQALLAKQAWRILHNPDALWVQILK 225
Query: 790 GVYFAQGDLLGAKRGYRPSYAWSSILQSSWIFQEGGRWKIGDGSQV 835
+YF D L + S+ WSS+L + + G+W++G G V
Sbjct: 226 ALYFPHHDFLQTTKRTGASWVWSSLLHGRELLLKQGQWQLGSGETV 363
>AV767121
Length = 525
Score = 88.2 bits (217), Expect = 7e-18
Identities = 55/175 (31%), Positives = 85/175 (48%), Gaps = 6/175 (3%)
Frame = +2
Query: 679 LKGWKESTLSRAGREVLIKSVVQAIPSYVMSCFILPDSLCADIERMVSRFYWGGDVDKRG 738
L WK+ T+S GR LI+SV+ A+P + +S F LP + R++ F WGG ++
Sbjct: 2 LSKWKQKTVSFGGRIFLIQSVLTALPLFFLSFFKLPIGVGKSCVRLMRNFLWGGSENENK 181
Query: 739 LQWASWKKLTRSKFDGGLGFRDFKSFNIALVAKNWWRIYTQPETLLAQVFKGVYFAQGDL 798
+ W + + K GGLG +D +FN AL+ K WR T+P++L +V + AQ D
Sbjct: 182 IA*VKWTDVCKPKELGGLGIKDLFTFNKALLGK*RWRYLTEPDSLWRRVIE----AQPDH 349
Query: 799 LGAKRGYRPSYAWSSILQ------SSWIFQEGGRWKIGDGSQVDILHDQWLPQGV 847
+ W+ IL W F G + +G+G Q + WL G+
Sbjct: 350 YSCGSSW-----WNDILSLCPEDADGW-FSSGLKKLVGEGDQTKFWSEDWLGSGL 496
>BP080049
Length = 356
Score = 86.7 bits (213), Expect = 2e-17
Identities = 43/102 (42%), Positives = 63/102 (61%)
Frame = -1
Query: 1158 LVFRWSMQLTIQMEFRRVLFETDCLQLFQLWKKPPDGRSYLSSIVGDCFMLSRFFDYVDL 1217
L RW++ L ++ FR ++FETDCL L++ WK+ G S L +++ DC L +FD L
Sbjct: 356 LSLRWALGLASELGFR*IVFETDCLHLYEAWKRR-GGCSVLDTLILDCRNLLSYFDVFQL 180
Query: 1218 SFVRRRGNSVADFLAGTASKYTDMVWLEEVPREVITLIHKDV 1259
SFVRR N V D LA + ++VW+EE+P E+ L+ DV
Sbjct: 179 SFVRRTSNCVVDALAKLCFQLGNVVWVEELPHELYGLVQSDV 54
>AV780250
Length = 494
Score = 86.3 bits (212), Expect = 3e-17
Identities = 43/110 (39%), Positives = 57/110 (51%)
Frame = +3
Query: 904 DTLMWPETVDGNYCSKSGYIFVRKNFLGACSSTYSQPSLPAPLWKKFWRTSAQPRCKEVA 963
D L WP T +G Y SKSGY VR + A S+ + + +WK PRC+E+
Sbjct: 174 DVLAWPHTSNGEYTSKSGYAVVRNKVVHALPSSSTALQVSPVVWK----APTLPRCRELI 341
Query: 964 WRVVSSLLPIRAALHRRGMDVDPLCPVCANEEETVFHLFVSCPVAQNFWF 1013
R +LP R+AL RGMDVD LCP C E+ + + PV +WF
Sbjct: 342 GRAFLKILPTRSALRARGMDVDELCPSCETHRESPAQVLLFVPVVSKWWF 491
>TC20198 similar to UP|Q9ZPU0 (Q9ZPU0) At2g13980 protein, partial (13%)
Length = 630
Score = 86.3 bits (212), Expect = 3e-17
Identities = 55/160 (34%), Positives = 80/160 (49%), Gaps = 2/160 (1%)
Frame = +3
Query: 1098 LLQATWARPPYGIYKLNFDAAVATTGEVGFGLIVRNMLGEVLASAAQYLLHAASAILGEA 1157
+L W PP G++KLNFD +V + G L+VR+ G + S + + G +
Sbjct: 78 MLAECWPPPPDGVFKLNFDTSVLASS*AGLSLVVRDCAG*EVFSIQ-------N*VFGAS 236
Query: 1158 LVFRW--SMQLTIQMEFRRVLFETDCLQLFQLWKKPPDGRSYLSSIVGDCFMLSRFFDYV 1215
L+ R S + + + V+ +TDC LF WKK SY F+L F +V
Sbjct: 237 LISRDG*SYSVLVDLFLTSVILKTDCYNLFVQWKKKHVSASYFGVYCRTFFLLCACFSFV 416
Query: 1216 DLSFVRRRGNSVADFLAGTASKYTDMVWLEEVPREVITLI 1255
DLSFV RRGN VA+ LA A VW+E+ P ++ L+
Sbjct: 417 DLSFV-RRGNKVANDLAKLAFSSHGYVWIEDSPIGLVALL 533
>AV411435
Length = 426
Score = 83.6 bits (205), Expect = 2e-16
Identities = 44/138 (31%), Positives = 64/138 (45%), Gaps = 2/138 (1%)
Frame = +3
Query: 892 ILKIHLPLNGSVDTLMWPETVDGNYCSKSGYIFVRKNF--LGACSSTYSQPSLPAPLWKK 949
I++ + GS D L WP G Y KSGY V+ L +S+ S PS + LW
Sbjct: 3 IMQTPFSITGSEDRLFWPFIPSGEYSVKSGYRAVKATQFNLSNLASSSSHPS--SFLWHT 176
Query: 950 FWRTSAQPRCKEVAWRVVSSLLPIRAALHRRGMDVDPLCPVCANEEETVFHLFVSCPVAQ 1009
W + + WR ++ +P++ L RR M D CP+C E + H ++C +
Sbjct: 177 IWGAQVPKKVRSFLWRAANNAIPVKRNLKRRNMGRDDFCPICNKGPEDINHALLACEWTR 356
Query: 1010 NFWFGSPLSLRVHGFSSM 1027
WFG PL+ VH M
Sbjct: 357 AVWFGCPLNFVVHEVHGM 410
>TC10764
Length = 555
Score = 71.6 bits (174), Expect = 7e-13
Identities = 38/106 (35%), Positives = 61/106 (56%)
Frame = +1
Query: 1115 FDAAVATTGEVGFGLIVRNMLGEVLASAAQYLLHAASAILGEALVFRWSMQLTIQMEFRR 1174
FD +V +VGFGL+ + GEVLA+ + + L ++ EA F W++ + F
Sbjct: 76 FDVSVVEHLDVGFGLLAGDHEGEVLAATSAFPLSVLPPLIAEA--FCWALSSDMTSCFGS 249
Query: 1175 VLFETDCLQLFQLWKKPPDGRSYLSSIVGDCFMLSRFFDYVDLSFV 1220
FE LQL++ W K G SY+ ++V DC +L+ +FD+++ SFV
Sbjct: 250 FCFEIGYLQLYETWMKT*PGSSYVCTLVNDCRLLACYFDFIEPSFV 387
>TC11350 similar to UP|Q9LJP7 (Q9LJP7) Genomic DNA, chromosome 3, P1 clone:
MIG10, partial (18%)
Length = 1118
Score = 71.6 bits (174), Expect = 7e-13
Identities = 58/194 (29%), Positives = 94/194 (47%), Gaps = 19/194 (9%)
Frame = -3
Query: 1012 WFGSPLSLRVHG---------FSSMEDFLADFFRAADDDALALWQAGVYALWEMRNRVVF 1062
WFGSPL + F F+ + ++D LA ++A+W+ RNRV+F
Sbjct: 1116 WFGSPLQWSIEPGGNLRFDRWFVERSQFILNHSLNHEED-LATLSFTLWAIWKNRNRVIF 940
Query: 1063 RDGEIPVPVAA------MIQRCSMLAAAPVSEMTVVPRPAPLLQA---TWARPPYGIYKL 1113
+ E P PV M Q ++++ + ++ T R + A +W PP G+YKL
Sbjct: 939 QLEE-PNPVNTIFQIRHMQQDANLMSTSQSNKQTTRERNINVTSARDRSWRPPPLGVYKL 763
Query: 1114 NFDAA-VATTGEVGFGLIVRNMLGEVLASAAQYLLHAASAILGEALVFRWSMQLTIQMEF 1172
N DA+ A + G+++R+ LG +++ A L A S I+ EAL R + L +
Sbjct: 762 NSDASWKAPSPSCTVGVVIRDNLGLLISGTASRPL-APSPIVSEALALREGLILARSLGL 586
Query: 1173 RRVLFETDCLQLFQ 1186
R+L E+DC L +
Sbjct: 585 DRILSESDCQPLIE 544
>TC17368
Length = 563
Score = 51.2 bits (121), Expect(2) = 7e-13
Identities = 27/69 (39%), Positives = 40/69 (57%)
Frame = +3
Query: 1160 FRWSMQLTIQMEFRRVLFETDCLQLFQLWKKPPDGRSYLSSIVGDCFMLSRFFDYVDLSF 1219
FRW+++LT+ + F V+FE DC L W + YLS IV D + LS F+ +
Sbjct: 228 FRWAIELTMDLGFSGVIFENDC*VLHCAWISKLNSPPYLSFIVQDYWFLSFGFNSCSFVY 407
Query: 1220 VRRRGNSVA 1228
V+R+ N+VA
Sbjct: 408 VKRQCNAVA 434
Score = 40.4 bits (93), Expect(2) = 7e-13
Identities = 20/55 (36%), Positives = 29/55 (52%)
Frame = +1
Query: 1103 WARPPYGIYKLNFDAAVATTGEVGFGLIVRNMLGEVLASAAQYLLHAASAILGEA 1157
W P G +NF A+ GFGLI RN G+VLA A ++ +S ++ +A
Sbjct: 55 WKPPAQGSINVNFGASFKAGVGAGFGLIARNGSGQVLAIATNLIVDVSSPVVAQA 219
>BP075797
Length = 504
Score = 70.5 bits (171), Expect = 2e-12
Identities = 46/129 (35%), Positives = 62/129 (47%)
Frame = -2
Query: 1130 IVRNMLGEVLASAAQYLLHAASAILGEALVFRWSMQLTIQMEFRRVLFETDCLQLFQLWK 1189
I N G+ + +++ +LL A + EAL R ++ L FR + E+DCLQL W
Sbjct: 473 IFYNHEGKAIIASSFHLLVAYEPAIAEALSLRRTITLPQDENFRIIELESDCLQLVHAWD 294
Query: 1190 KPPDGRSYLSSIVGDCFMLSRFFDYVDLSFVRRRGNSVADFLAGTASKYTDMVWLEEVPR 1249
+ YL+SIV DC LS F D L R+ N VAD LA A Y D W+ P
Sbjct: 293 RSVPHPHYLASIVQDCKDLSSFSDSCSLKHSSRKINEVADHLAKLAFVYQDHFWVGIDPP 114
Query: 1250 EVITLIHKD 1258
L+ D
Sbjct: 113 ATSLLLQPD 87
>TC12829
Length = 448
Score = 70.1 bits (170), Expect = 2e-12
Identities = 35/60 (58%), Positives = 43/60 (71%), Gaps = 2/60 (3%)
Frame = +3
Query: 684 ESTLSRAGREVLIKSVVQAIPSYVMSCFILPDSLCADIERMVSRFY--WGGDVDKRGLQW 741
E L RAGREVLIKSV +AIP+Y+MSCF LPDS+C+ IE MV +FY W ++ L W
Sbjct: 252 EKFLYRAGREVLIKSVTKAIPTYIMSCFALPDSICSQIEGMVGKFYLEW*CVSEEYSLAW 431
Score = 33.9 bits (76), Expect = 0.16
Identities = 11/20 (55%), Positives = 13/20 (65%)
Frame = +1
Query: 728 FYWGGDVDKRGLQWASWKKL 747
F W GDV +R + W WKKL
Sbjct: 385 FIWSGDVSRRSIHWLGWKKL 444
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.324 0.139 0.438
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 25,026,132
Number of Sequences: 28460
Number of extensions: 391001
Number of successful extensions: 2599
Number of sequences better than 10.0: 102
Number of HSP's better than 10.0 without gapping: 2548
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2578
length of query: 1259
length of database: 4,897,600
effective HSP length: 101
effective length of query: 1158
effective length of database: 2,023,140
effective search space: 2342796120
effective search space used: 2342796120
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.5 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0129.4


