

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0100a.10
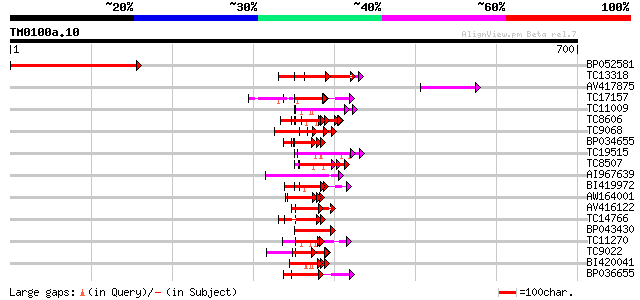
(700 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BP052581 333 6e-92
TC13318 similar to UP|CAA20314 (CAA20314) SPBC30B4.01c protein (... 65 3e-11
AV417875 63 2e-10
TC17157 similar to UP|CAD27462 (CAD27462) Nucleosome assembly pr... 62 4e-10
TC11009 similar to GB|AAP21377.1|30102918|BT006569 At1g47970 {Ar... 60 1e-09
TC8606 similar to UP|AAQ82688 (AAQ82688) Epa4p, partial (7%) 58 4e-09
TC9068 similar to UP|SCWA_YEAST (Q04951) Probable family 17 gluc... 56 2e-08
BP034655 54 1e-07
TC19515 similar to UP|Q9YGK0 (Q9YGK0) Vitellogenin precursor, pa... 52 2e-07
TC8507 similar to UP|Q8LJS2 (Q8LJS2) Nucleolar histone deacetyla... 50 9e-07
AI967639 50 1e-06
BI419972 49 2e-06
AW164001 47 8e-06
AV416122 47 1e-05
TC14766 similar to UP|Q7XXR8 (Q7XXR8) Nascent polypeptide associ... 46 2e-05
BP043430 46 2e-05
TC11270 similar to UP|Q9LM88 (Q9LM88) F2D10.15, partial (9%) 45 3e-05
TC9022 similar to PIR|T46225|T46225 alpha NAC-like protein - Ara... 45 3e-05
BI420041 45 4e-05
BP036655 45 5e-05
>BP052581
Length = 563
Score = 333 bits (854), Expect = 6e-92
Identities = 162/162 (100%), Positives = 162/162 (100%)
Frame = +1
Query: 1 MASMSGPTAPNLTFPNKNLSIPFTLKRPSLLPFPSHPPCRCLSSASDDKNDSSDNRRWDS 60
MASMSGPTAPNLTFPNKNLSIPFTLKRPSLLPFPSHPPCRCLSSASDDKNDSSDNRRWDS
Sbjct: 76 MASMSGPTAPNLTFPNKNLSIPFTLKRPSLLPFPSHPPCRCLSSASDDKNDSSDNRRWDS 255
Query: 61 LLQDFLTGAAKQFDSFMNALRDGRSAEDRAATAGGGSDEDWDWDRWRQHFDEVDDQERLL 120
LLQDFLTGAAKQFDSFMNALRDGRSAEDRAATAGGGSDEDWDWDRWRQHFDEVDDQERLL
Sbjct: 256 LLQDFLTGAAKQFDSFMNALRDGRSAEDRAATAGGGSDEDWDWDRWRQHFDEVDDQERLL 435
Query: 121 SVLKSQLGRAVYVEDYEDAARLKVAIAAASNNDSVGRVMSHL 162
SVLKSQLGRAVYVEDYEDAARLKVAIAAASNNDSVGRVMSHL
Sbjct: 436 SVLKSQLGRAVYVEDYEDAARLKVAIAAASNNDSVGRVMSHL 561
>TC13318 similar to UP|CAA20314 (CAA20314) SPBC30B4.01c protein (SPBC3D6.14C
protein) (Fragment), partial (15%)
Length = 543
Score = 65.5 bits (158), Expect = 3e-11
Identities = 24/45 (53%), Positives = 40/45 (88%)
Frame = +3
Query: 352 DEEDDEDEDEDEDEDEDEDDDDDEDEDEDEDEDEEKDSDTGSLEL 396
+EEDD++E++DEDED+D+DDDDD+D +E+ED+D++ + + GS E+
Sbjct: 255 EEEDDDEEEDDEDEDDDDDDDDDDDGEEEEDDDDDDEEEEGSEEV 389
Score = 63.5 bits (153), Expect = 1e-10
Identities = 34/96 (35%), Positives = 58/96 (60%), Gaps = 1/96 (1%)
Frame = +3
Query: 333 ITPEKVDKDLISKVIEQIIDEEDDEDEDEDEDEDEDEDDDDDEDEDEDEDEDEEKDSDTG 392
I P+ K + + E+ D+E+++DEDED+D+D+D+DDD +E+ED+D+D++EE+ S+
Sbjct: 210 IYPDGPPKPVQTDNEEEEDDDEEEDDEDEDDDDDDDDDDDGEEEEDDDDDDEEEEGSEEV 389
Query: 393 SLELVD-MKSETDQEADDEVEINTGLGTSDREDQNE 427
+E D K Q + G+G+ D NE
Sbjct: 390 EVEGKDGAKGAAAQTTKAAAAMCVGIGSF--SDPNE 491
Score = 53.9 bits (128), Expect = 8e-08
Identities = 24/74 (32%), Positives = 42/74 (56%)
Frame = +3
Query: 364 DEDEDEDDDDDEDEDEDEDEDEEKDSDTGSLELVDMKSETDQEADDEVEINTGLGTSDRE 423
D +E+EDDD++ED+++++D+D++ D D G E D + ++E +EVE+ G
Sbjct: 246 DNEEEEDDDEEEDDEDEDDDDDDDDDDDGEEEEDDDDDDEEEEGSEEVEVEGKDGAKGAA 425
Query: 424 DQNEIAVKVVIGGI 437
Q A + GI
Sbjct: 426 AQTTKAAAAMCVGI 467
>AV417875
Length = 314
Score = 62.8 bits (151), Expect = 2e-10
Identities = 33/76 (43%), Positives = 45/76 (58%), Gaps = 2/76 (2%)
Frame = +3
Query: 508 FIGKGKVPAKVLKEVGELINLTLSQAQNHQPLSGSTIFNRIEIPTS-FDPLNGLYIGVHG 566
F KV +KV K + E++ L +SQAQ L+ T F+RI FDP +GLY+G G
Sbjct: 78 FWNSDKVSSKVSKSIREILKLAVSQAQKKSRLTEDTCFSRITSSRGDFDPFDGLYVGAFG 257
Query: 567 LHS-EVIQLRRGFGQW 581
H E++QL+R FG W
Sbjct: 258 PHGIEIVQLKRKFGHW 305
>TC17157 similar to UP|CAD27462 (CAD27462) Nucleosome assembly protein
1-like protein 3, partial (81%)
Length = 1298
Score = 61.6 bits (148), Expect = 4e-10
Identities = 25/41 (60%), Positives = 34/41 (81%)
Frame = +2
Query: 352 DEEDDEDEDEDEDEDEDEDDDDDEDEDEDEDEDEEKDSDTG 392
D ED E++DED DEDEDEDDDDDE+E++D+D+DE+ + G
Sbjct: 758 DFEDIEEDDEDGDEDEDEDDDDDEEEEDDDDDDEDDEEGEG 880
Score = 60.8 bits (146), Expect = 7e-10
Identities = 41/103 (39%), Positives = 58/103 (55%), Gaps = 4/103 (3%)
Frame = +2
Query: 295 PESGDDRNDGSDPAERMPGFQNGLKDMIPGVKVKIF----KVITPEKVDKDLISKVIEQI 350
PE DD +D D E + D+ ++ KI T E D + IE+
Sbjct: 614 PEDDDDIDD--DAVEELQNLMEHDYDIGSTIRDKIIPHAVSWFTGEAEQSDF--EDIEED 781
Query: 351 IDEEDDEDEDEDEDEDEDEDDDDDEDEDEDEDEDEEKDSDTGS 393
DE+ DEDEDED+D+DE+E+DDDD+DED++E E + K S +GS
Sbjct: 782 -DEDGDEDEDEDDDDDEEEEDDDDDDEDDEEGEGKSK-SKSGS 904
Score = 45.4 bits (106), Expect = 3e-05
Identities = 24/94 (25%), Positives = 51/94 (53%), Gaps = 7/94 (7%)
Frame = +2
Query: 339 DKDLISKVIEQIIDE-------EDDEDEDEDEDEDEDEDDDDDEDEDEDEDEDEEKDSDT 391
D D+ S + ++II E ++ + ED +ED+++ D+D++++D+D++E+E+ D D
Sbjct: 677 DYDIGSTIRDKIIPHAVSWFTGEAEQSDFEDIEEDDEDGDEDEDEDDDDDEEEEDDDDD- 853
Query: 392 GSLELVDMKSETDQEADDEVEINTGLGTSDREDQ 425
E D+E + + + +G +DQ
Sbjct: 854 ---------DEDDEEGEGKSKSKSGSKARPGKDQ 928
>TC11009 similar to GB|AAP21377.1|30102918|BT006569 At1g47970 {Arabidopsis
thaliana;}, partial (44%)
Length = 700
Score = 60.1 bits (144), Expect = 1e-09
Identities = 34/97 (35%), Positives = 52/97 (53%), Gaps = 19/97 (19%)
Frame = +2
Query: 352 DEEDDED-----EDEDEDEDEDE--------------DDDDDEDEDEDEDEDEEKDSDTG 392
DE+DD+D +D+D++EDE+E DDDDD+D+D+DE+E+EE+D T
Sbjct: 65 DEDDDDDAPGGGDDDDDEEDEEEEGGVEGGRGGGGDPDDDDDDDDDDDEEEEEEEDLGTE 244
Query: 393 SLELVDMKSETDQEADDEVEINTGLGTSDREDQNEIA 429
L + + D+EA + E G + D E A
Sbjct: 245 YLVRRTVAAAEDEEASSDFEPEEDEGDDNDNDDGEKA 355
Score = 57.8 bits (138), Expect = 6e-09
Identities = 29/72 (40%), Positives = 43/72 (59%), Gaps = 5/72 (6%)
Frame = +2
Query: 353 EEDDEDEDEDEDEDEDEDDD-----DDEDEDEDEDEDEEKDSDTGSLELVDMKSETDQEA 407
+EDD+D+ ED+D DED+DDD DD+D++EDE+E+ + G D + D +
Sbjct: 26 DEDDDDDGEDDDGDEDDDDDAPGGGDDDDDEEDEEEEGGVEGGRGGGGDPDDDDDDDDDD 205
Query: 408 DDEVEINTGLGT 419
D+E E LGT
Sbjct: 206 DEEEEEEEDLGT 241
Score = 40.4 bits (93), Expect = 0.001
Identities = 24/81 (29%), Positives = 40/81 (48%), Gaps = 23/81 (28%)
Frame = +2
Query: 352 DEEDDEDEDEDEDEDEDED---------------DDD-------DEDEDEDEDEDE-EKD 388
D++DD+D+D+DE+E+E+ED D++ +EDE +D D D+ EK
Sbjct: 176 DDDDDDDDDDDEEEEEEEDLGTEYLVRRTVAAAEDEEASSDFEPEEDEGDDNDNDDGEKA 355
Query: 389 SDTGSLELVDMKSETDQEADD 409
+ D D ++DD
Sbjct: 356 GVPSKRKRSDKDGSGDDDSDD 418
Score = 36.2 bits (82), Expect = 0.018
Identities = 19/58 (32%), Positives = 29/58 (49%), Gaps = 7/58 (12%)
Frame = +2
Query: 342 LISKVIEQIIDEEDDEDEDEDEDEDEDEDDDDDED-------EDEDEDEDEEKDSDTG 392
L+ + + DEE D + +EDE +D D+DD E + D+D + DSD G
Sbjct: 248 LVRRTVAAAEDEEASSDFEPEEDEGDDNDNDDGEKAGVPSKRKRSDKDGSGDDDSDDG 421
Score = 33.9 bits (76), Expect = 0.090
Identities = 27/103 (26%), Positives = 43/103 (41%), Gaps = 9/103 (8%)
Frame = +2
Query: 294 DPESGDDRNDGSDPAERMPGFQNGLKDMIPGVKVKIFKVITPEKVDKDLISKVIEQIIDE 353
DP+ DD +D D E +D+ G + + + + + ++ S E DE
Sbjct: 170 DPDDDDDDDDDDDEEEEEE------EDL--GTEYLVRRTVAAAEDEE--ASSDFEPEEDE 319
Query: 354 EDDEDEDEDED---------EDEDEDDDDDEDEDEDEDEDEEK 387
DD D D+ E D+D DDD D+ ++DE K
Sbjct: 320 GDDNDNDDGEKAGVPSKRKRSDKDGSGDDDSDDGGEDDERPSK 448
>TC8606 similar to UP|AAQ82688 (AAQ82688) Epa4p, partial (7%)
Length = 1008
Score = 58.2 bits (139), Expect = 4e-09
Identities = 27/60 (45%), Positives = 44/60 (73%), Gaps = 2/60 (3%)
Frame = -2
Query: 353 EEDDEDEDEDEDEDEDEDDDDDEDEDEDED--EDEEKDSDTGSLELVDMKSETDQEADDE 410
EE+ EDE++++ ED+++DDDDDED+DE++D EDEE++ +E D + E + E D+E
Sbjct: 443 EENGEDEEDEDGEDQEDDDDDDEDDDEEDDGGEDEEEE----GVEEEDNEDEEEDEEDEE 276
Score = 55.8 bits (133), Expect = 2e-08
Identities = 24/56 (42%), Positives = 40/56 (70%)
Frame = -2
Query: 352 DEEDDEDEDEDEDEDEDEDDDDDEDEDEDEDEDEEKDSDTGSLELVDMKSETDQEA 407
DEED++ ED+++D+D+DEDDD+++D EDE+E+ ++ D E + E D+EA
Sbjct: 428 DEEDEDGEDQEDDDDDDEDDDEEDDGGEDEEEEGVEEEDNEDEE----EDEEDEEA 273
Score = 50.4 bits (119), Expect = 9e-07
Identities = 18/43 (41%), Positives = 35/43 (80%)
Frame = -2
Query: 348 EQIIDEEDDEDEDEDEDEDEDEDDDDDEDEDEDEDEDEEKDSD 390
+Q D++DDED+DE++D EDE+++ E+ED +++E++E+D +
Sbjct: 404 DQEDDDDDDEDDDEEDDGGEDEEEEGVEEEDNEDEEEDEEDEE 276
Score = 49.3 bits (116), Expect = 2e-06
Identities = 17/44 (38%), Positives = 35/44 (78%)
Frame = -2
Query: 352 DEEDDEDEDEDEDEDEDEDDDDDEDEDEDEDEDEEKDSDTGSLE 395
+++DD+DED+DE++D ED++++ E+ED +++EE + D +L+
Sbjct: 398 EDDDDDDEDDDEEDDGGEDEEEEGVEEEDNEDEEEDEEDEEALQ 267
Score = 48.9 bits (115), Expect = 3e-06
Identities = 24/48 (50%), Positives = 35/48 (72%), Gaps = 5/48 (10%)
Frame = -2
Query: 348 EQIIDEEDDEDEDEDEDEDEDEDDDDDEDE-----DEDEDEDEEKDSD 390
E+ D ED ED D+D+DED+DE+DD EDE +E+++EDEE+D +
Sbjct: 425 EEDEDGEDQED-DDDDDEDDDEEDDGGEDEEEEGVEEEDNEDEEEDEE 285
Score = 48.5 bits (114), Expect = 4e-06
Identities = 19/52 (36%), Positives = 36/52 (68%)
Frame = -2
Query: 361 EDEDEDEDEDDDDDEDEDEDEDEDEEKDSDTGSLELVDMKSETDQEADDEVE 412
E+ EDE+++D +D+++D+D+DED++++ D G E + E D E ++E E
Sbjct: 443 EENGEDEEDEDGEDQEDDDDDDEDDDEEDDGGEDEEEEGVEEEDNEDEEEDE 288
Score = 45.4 bits (106), Expect = 3e-05
Identities = 23/57 (40%), Positives = 41/57 (71%), Gaps = 4/57 (7%)
Frame = -2
Query: 335 PEKVDKDLISKVIEQIIDEEDDEDEDEDED---EDEDEDDDDDED-EDEDEDEDEEK 387
PE+ +D + E D++DD+++D++ED EDE+E+ ++ED EDE+EDE++E+
Sbjct: 446 PEENGEDEEDEDGEDQEDDDDDDEDDDEEDDGGEDEEEEGVEEEDNEDEEEDEEDEE 276
Score = 40.0 bits (92), Expect = 0.001
Identities = 17/38 (44%), Positives = 29/38 (75%), Gaps = 2/38 (5%)
Frame = -2
Query: 352 DEEDDEDEDEDED--EDEDEDDDDDEDEDEDEDEDEEK 387
DEEDD EDE+E+ E+ED +D+++++EDE+ + +K
Sbjct: 368 DEEDDGGEDEEEEGVEEEDNEDEEEDEEDEEALQPPKK 255
Score = 36.6 bits (83), Expect = 0.014
Identities = 30/112 (26%), Positives = 42/112 (36%), Gaps = 36/112 (32%)
Frame = -2
Query: 352 DEEDDEDEDEDEDEDEDEDD----------------------------------DDDEDE 377
D+E DED+D D E E+D ++ EDE
Sbjct: 602 DDEGDEDDDGDGPFGEGEEDLSSEDGAGYGNNSNNKSNSKKAPEGGPGAGAGPEENGEDE 423
Query: 378 DEDEDEDEEKDSDTGSLELVDMKSETDQEADD--EVEINTGLGTSDREDQNE 427
++++ ED+E D D E D E DD E E G+ D ED+ E
Sbjct: 422 EDEDGEDQEDDDD---------DDEDDDEEDDGGEDEEEEGVEEEDNEDEEE 294
>TC9068 similar to UP|SCWA_YEAST (Q04951) Probable family 17 glucosidase
SCW10 precursor (Soluble cell wall protein 10) ,
partial (6%)
Length = 1209
Score = 56.2 bits (134), Expect = 2e-08
Identities = 19/36 (52%), Positives = 33/36 (90%)
Frame = +2
Query: 358 DEDEDEDEDEDEDDDDDEDEDEDEDEDEEKDSDTGS 393
D+D+D+D+D+D+DDDDD+D+D+D+ + ++ DSDTG+
Sbjct: 83 DDDDDDDDDDDDDDDDDDDDDDDDGDKKQPDSDTGT 190
Score = 44.7 bits (104), Expect = 5e-05
Identities = 19/53 (35%), Positives = 33/53 (61%)
Frame = +2
Query: 328 KIFKVITPEKVDKDLISKVIEQIIDEEDDEDEDEDEDEDEDEDDDDDEDEDED 380
++ K+ T K L S + D++DD+D+D+D+D+D+D+DD D + D D
Sbjct: 23 QLTKINTAPSGFKRLFSPKPDDDDDDDDDDDDDDDDDDDDDDDDGDKKQPDSD 181
Score = 43.5 bits (101), Expect = 1e-04
Identities = 16/36 (44%), Positives = 26/36 (71%)
Frame = +2
Query: 368 DEDDDDDEDEDEDEDEDEEKDSDTGSLELVDMKSET 403
D+DDDDD+D+D+D+D+D++ D D G + D + T
Sbjct: 83 DDDDDDDDDDDDDDDDDDDDDDDDGDKKQPDSDTGT 190
>BP034655
Length = 517
Score = 53.5 bits (127), Expect = 1e-07
Identities = 16/39 (41%), Positives = 38/39 (97%)
Frame = +1
Query: 352 DEEDDEDEDEDEDEDEDEDDDDDEDEDEDEDEDEEKDSD 390
D+E++E+E+E+E+E+E+E+++++E+E+++ED++E++D D
Sbjct: 139 DDEEEEEEEEEEEEEEEEEEEEEEEEEDEEDDEEDEDED 255
Score = 53.5 bits (127), Expect = 1e-07
Identities = 16/40 (40%), Positives = 38/40 (95%)
Frame = +1
Query: 351 IDEEDDEDEDEDEDEDEDEDDDDDEDEDEDEDEDEEKDSD 390
+ ++D+E+E+E+E+E+E+E+++++E+E+E+++ED+E+D D
Sbjct: 130 LSDDDEEEEEEEEEEEEEEEEEEEEEEEEEDEEDDEEDED 249
Score = 53.1 bits (126), Expect = 1e-07
Identities = 19/37 (51%), Positives = 35/37 (94%)
Frame = +1
Query: 348 EQIIDEEDDEDEDEDEDEDEDEDDDDDEDEDEDEDED 384
E+ +EE++E+E+E+E+E+E+E+++D+ED++EDEDED
Sbjct: 145 EEEEEEEEEEEEEEEEEEEEEEEEEDEEDDEEDEDED 255
Score = 52.4 bits (124), Expect = 2e-07
Identities = 16/39 (41%), Positives = 37/39 (94%)
Frame = +1
Query: 352 DEEDDEDEDEDEDEDEDEDDDDDEDEDEDEDEDEEKDSD 390
D++++E+E+E+E+E+E+E+++++E+E+EDE++DEE + +
Sbjct: 136 DDDEEEEEEEEEEEEEEEEEEEEEEEEEDEEDDEEDEDE 252
Score = 45.4 bits (106), Expect = 3e-05
Identities = 18/40 (45%), Positives = 32/40 (80%)
Frame = +1
Query: 339 DKDLISKVIEQIIDEEDDEDEDEDEDEDEDEDDDDDEDED 378
D D + E+ +EE++E+E+E+E+E+++EDD++DEDED
Sbjct: 136 DDDEEEEEEEEEEEEEEEEEEEEEEEEEDEEDDEEDEDED 255
Score = 39.3 bits (90), Expect = 0.002
Identities = 24/80 (30%), Positives = 39/80 (48%), Gaps = 2/80 (2%)
Frame = +1
Query: 372 DDDEDEDEDEDEDEEKDSDTGSLELVDMKSETDQEADDE--VEINTGLGTSDREDQNEIA 429
DDDE+E+E+E+E+EE++ + E + E D+E +DE + + S QN +
Sbjct: 136 DDDEEEEEEEEEEEEEEEEEEEEEEEEEDEEDDEEDEDEDYIPVAPSHRVSPNVHQNPTS 315
Query: 430 VKVVIGGIVQKLSSNISTRD 449
V + I N S D
Sbjct: 316 DSVEVILIPDSEQQNYSEED 375
>TC19515 similar to UP|Q9YGK0 (Q9YGK0) Vitellogenin precursor, partial (3%)
Length = 504
Score = 52.4 bits (124), Expect = 2e-07
Identities = 32/86 (37%), Positives = 48/86 (55%), Gaps = 14/86 (16%)
Frame = +3
Query: 356 DEDEDEDEDEDEDEDDDDDEDEDEDEDE------DEEKDSDTGSLELVDMKSETDQE-AD 408
D+D D D+ E EDE+DDDD+D+ +DED E DSD S + D S+ D E AD
Sbjct: 57 DDDSDPDDAESEDEEDDDDDDDGDDEDPLLGPGLGRESDSDDMSHDDYDSASDIDDEDAD 236
Query: 409 ---DEVEINTGLG----TSDREDQNE 427
D+++ + G G DR++ ++
Sbjct: 237 FILDDLDFDGGTGLLEIVGDRDEDDD 314
Score = 49.7 bits (117), Expect = 2e-06
Identities = 34/117 (29%), Positives = 52/117 (44%), Gaps = 30/117 (25%)
Frame = +3
Query: 352 DEEDDEDEDEDEDEDEDEDDDDDED---------------------------EDEDED-- 382
D + D+ E EDE++D+D+DD DDED +DED D
Sbjct: 63 DSDPDDAESEDEEDDDDDDDGDDEDPLLGPGLGRESDSDDMSHDDYDSASDIDDEDADFI 242
Query: 383 -EDEEKDSDTGSLELVDMKSETDQEADDEVEINTGLGTSDREDQNEIAVKVVIGGIV 438
+D + D TG LE+V + D++ DD E+ + + D N+ + I G V
Sbjct: 243 LDDLDFDGGTGLLEIV---GDRDEDDDDSQELESASSDDEDFDDNDFGY*LSI*GTV 404
Score = 35.8 bits (81), Expect = 0.024
Identities = 29/109 (26%), Positives = 48/109 (43%), Gaps = 11/109 (10%)
Frame = +3
Query: 281 TEDRSELFVVSTEDPESGDDRNDGSDPAERM-PGF--QNGLKDMIPGVKVKIFKVITPEK 337
T+D S+ +ED E DD +DG D + PG ++ DM + +
Sbjct: 54 TDDDSDPDDAESEDEEDDDDDDDGDDEDPLLGPGLGRESDSDDMSHDDYDSASDI---DD 224
Query: 338 VDKDLISKVIE--------QIIDEEDDEDEDEDEDEDEDEDDDDDEDED 378
D D I ++ +I+ + D++D+D E E DD+D +D D
Sbjct: 225 EDADFILDDLDFDGGTGLLEIVGDRDEDDDDSQELESASSDDEDFDDND 371
>TC8507 similar to UP|Q8LJS2 (Q8LJS2) Nucleolar histone deacetylase
HD2-P39, partial (40%)
Length = 620
Score = 50.4 bits (119), Expect = 9e-07
Identities = 26/58 (44%), Positives = 35/58 (59%), Gaps = 6/58 (10%)
Frame = +3
Query: 352 DEEDDEDEDEDEDEDEDEDDDD---DEDEDEDEDEDEE---KDSDTGSLELVDMKSET 403
DEEDD D+++DED+D DE+ DD D DE + +D DEE K +D G D +T
Sbjct: 3 DEEDDSDDEDDEDDDSDEEMDDADSDSDESDSDDTDEETPVKKADQGKKRANDSALKT 176
Score = 48.5 bits (114), Expect = 4e-06
Identities = 20/62 (32%), Positives = 38/62 (61%)
Frame = +3
Query: 358 DEDEDEDEDEDEDDDDDEDEDEDEDEDEEKDSDTGSLELVDMKSETDQEADDEVEINTGL 417
DE++D D+++DEDDD DE+ D+ + + +E DSD E K++ ++ ++ + T +
Sbjct: 3 DEEDDSDDEDDEDDDSDEEMDDADSDSDESDSDDTDEETPVKKADQGKKRANDSALKTPV 182
Query: 418 GT 419
T
Sbjct: 183 ST 188
Score = 46.2 bits (108), Expect = 2e-05
Identities = 19/53 (35%), Positives = 31/53 (57%)
Frame = +3
Query: 357 EDEDEDEDEDEDEDDDDDEDEDEDEDEDEEKDSDTGSLELVDMKSETDQEADD 409
++ED+ +DED+++DD D+E +D D D DE DT V + + A+D
Sbjct: 3 DEEDDSDDEDDEDDDSDEEMDDADSDSDESDSDDTDEETPVKKADQGKKRAND 161
Score = 37.0 bits (84), Expect = 0.011
Identities = 28/115 (24%), Positives = 53/115 (45%), Gaps = 2/115 (1%)
Frame = +3
Query: 377 EDEDEDEDEEKDSDTGSLELVDMKSETDQEADDEVEINTGLGTSD--REDQNEIAVKVVI 434
++ED+ +DE+ + D E+ D S++D+ D+ + T + +D ++ N+ A+K +
Sbjct: 3 DEEDDSDDEDDEDDDSDEEMDDADSDSDESDSDDTDEETPVKKADQGKKRANDSALKTPV 182
Query: 435 GGIVQKLSSNISTRDLLRVPAKLEIKKRGSFSFTVEKEVNQQDGQAKGKSSSDKS 489
K + NI+ P K + KK G G+ GK+S K+
Sbjct: 183 S---TKKAKNIT-------PEKTDGKKTG-------HTATPHPGKKGGKNSESKA 296
>AI967639
Length = 335
Score = 50.1 bits (118), Expect = 1e-06
Identities = 34/97 (35%), Positives = 49/97 (50%), Gaps = 1/97 (1%)
Frame = +1
Query: 317 GLKDMIPGVKVKIFKVITPEKVDKDLISKVIEQIIDEEDDE-DEDEDEDEDEDEDDDDDE 375
G D I G V + + K +D+ S DEE+D+ DE EDE+ED+DDDDD+
Sbjct: 7 GTSDDIVGEVVYDEEYLKKRKQRRDISSSSEGDEEDEEEDQGDEYNLEDEEEDDDDDDDD 186
Query: 376 DEDEDEDEDEEKDSDTGSLELVDMKSETDQEADDEVE 412
D ED D +K L + ET + + DE++
Sbjct: 187 DLSISEDSDSDKPRKVKQLP-GRTRRETKRRSVDEIQ 294
Score = 30.8 bits (68), Expect = 0.76
Identities = 22/79 (27%), Positives = 34/79 (42%), Gaps = 12/79 (15%)
Frame = +1
Query: 348 EQIIDEE---------DDEDEDEDEDEDEDEDDDDD---EDEDEDEDEDEEKDSDTGSLE 395
E + DEE D E ++EDE+ED D+ EDE+ED+D+D+
Sbjct: 31 EVVYDEEYLKKRKQRRDISSSSEGDEEDEEEDQGDEYNLEDEEEDDDDDD---------- 180
Query: 396 LVDMKSETDQEADDEVEIN 414
DD++ I+
Sbjct: 181 ------------DDDLSIS 201
Score = 28.5 bits (62), Expect = 3.8
Identities = 17/68 (25%), Positives = 32/68 (47%)
Frame = +1
Query: 360 DEDEDEDEDEDDDDDEDEDEDEDEDEEKDSDTGSLELVDMKSETDQEADDEVEINTGLGT 419
DE+ + + D + DE+++EE D +LE D+E DD+ + + L
Sbjct: 43 DEEYLKKRKQRRDISSSSEGDEEDEEEDQGDEYNLE--------DEEEDDDDDDDDDLSI 198
Query: 420 SDREDQNE 427
S+ D ++
Sbjct: 199 SEDSDSDK 222
>BI419972
Length = 498
Score = 49.3 bits (116), Expect = 2e-06
Identities = 20/39 (51%), Positives = 28/39 (71%)
Frame = +1
Query: 352 DEEDDEDEDEDEDEDEDEDDDDDEDEDEDEDEDEEKDSD 390
D E ++DED D+++D+ +DDDDDE ED DE EE D +
Sbjct: 343 DSETEDDEDGDDEDDDADDDDDDEGEDYSGDEGEEADPE 459
Score = 48.1 bits (113), Expect = 5e-06
Identities = 18/51 (35%), Positives = 36/51 (70%)
Frame = +1
Query: 340 KDLISKVIEQIIDEEDDEDEDEDEDEDEDEDDDDDEDEDEDEDEDEEKDSD 390
+DL+ + + +++ + ++DED D+++D+ DDDD+DE ED DE +++D
Sbjct: 301 EDLLGRKPAYLANKDSETEDDEDGDDEDDDADDDDDDEGEDYSGDEGEEAD 453
Score = 45.8 bits (107), Expect = 2e-05
Identities = 24/64 (37%), Positives = 35/64 (54%)
Frame = +1
Query: 358 DEDEDEDEDEDEDDDDDEDEDEDEDEDEEKDSDTGSLELVDMKSETDQEADDEVEINTGL 417
++D + ++DED DD+DD+ +D+D+DE E+ D G E D E D E G
Sbjct: 337 NKDSETEDDEDGDDEDDDADDDDDDEGEDYSGDEG--------EEADPEDDPEA---NGA 483
Query: 418 GTSD 421
G SD
Sbjct: 484 GGSD 495
Score = 42.7 bits (99), Expect = 2e-04
Identities = 20/45 (44%), Positives = 29/45 (64%), Gaps = 3/45 (6%)
Frame = +1
Query: 352 DEEDDEDEDED---EDEDEDEDDDDDEDEDEDEDEDEEKDSDTGS 393
D+ED +DED+D +D+DE ED DE E+ D ++D E + GS
Sbjct: 358 DDEDGDDEDDDADDDDDDEGEDYSGDEGEEADPEDDPEANGAGGS 492
Score = 31.6 bits (70), Expect = 0.45
Identities = 13/34 (38%), Positives = 20/34 (58%)
Frame = +1
Query: 352 DEEDDEDEDEDEDEDEDEDDDDDEDEDEDEDEDE 385
D++DDE ED DE E+ D +DD + + D+
Sbjct: 397 DDDDDEGEDYSGDEGEEADPEDDPEANGAGGSDD 498
>AW164001
Length = 447
Score = 47.4 bits (111), Expect = 8e-06
Identities = 17/41 (41%), Positives = 33/41 (80%)
Frame = +1
Query: 344 SKVIEQIIDEEDDEDEDEDEDEDEDEDDDDDEDEDEDEDED 384
S V+ +++++D++ ++ED DED DDDDD+D+D+D+D++
Sbjct: 13 SDVVGDSVNKQNDDEVYKEEDVDEDFDDDDDDDDDDDDDDE 135
Score = 44.7 bits (104), Expect = 5e-05
Identities = 17/40 (42%), Positives = 30/40 (74%)
Frame = +1
Query: 341 DLISKVIEQIIDEEDDEDEDEDEDEDEDEDDDDDEDEDED 380
D++ + + D+E ++ED DED D+D+DDDDD+D+D++
Sbjct: 16 DVVGDSVNKQNDDEVYKEEDVDEDFDDDDDDDDDDDDDDE 135
Score = 43.5 bits (101), Expect = 1e-04
Identities = 19/48 (39%), Positives = 32/48 (66%)
Frame = +1
Query: 341 DLISKVIEQIIDEEDDEDEDEDEDEDEDEDDDDDEDEDEDEDEDEEKD 388
D ++K + + +E+D DED D+D+D+D+DDDDD++ D+ KD
Sbjct: 28 DSVNKQNDDEVYKEEDVDEDFDDDDDDDDDDDDDDEPATLGFVDKPKD 171
Score = 32.7 bits (73), Expect = 0.20
Identities = 12/39 (30%), Positives = 27/39 (68%)
Frame = +1
Query: 333 ITPEKVDKDLISKVIEQIIDEEDDEDEDEDEDEDEDEDD 371
+ + V+K +V ++ +ED +D+D+D+D+D+D+D+
Sbjct: 19 VVGDSVNKQNDDEVYKEEDVDEDFDDDDDDDDDDDDDDE 135
Score = 30.4 bits (67), Expect = 1.00
Identities = 13/36 (36%), Positives = 25/36 (69%), Gaps = 2/36 (5%)
Frame = +1
Query: 348 EQIIDEE--DDEDEDEDEDEDEDEDDDDDEDEDEDE 381
E+ +DE+ DD+D+D+D+D+D++ D+ +DE
Sbjct: 67 EEDVDEDFDDDDDDDDDDDDDDEPATLGFVDKPKDE 174
>AV416122
Length = 414
Score = 47.0 bits (110), Expect = 1e-05
Identities = 20/49 (40%), Positives = 35/49 (70%)
Frame = +3
Query: 354 EDDEDEDEDEDEDEDEDDDDDEDEDEDEDEDEEKDSDTGSLELVDMKSE 402
+D+E +++ + E E+E+DD+++ E+EDE+ED+E D G E VD K +
Sbjct: 264 DDNEPDNDVQFEGEEEEDDEEDKEEEDEEEDDEDDDSNGEAE-VDRKGK 407
Score = 42.0 bits (97), Expect = 3e-04
Identities = 17/39 (43%), Positives = 28/39 (71%)
Frame = +3
Query: 349 QIIDEEDDEDEDEDEDEDEDEDDDDDEDEDEDEDEDEEK 387
Q EE+++DE++ E+EDE+EDD+DD+ E E + + K
Sbjct: 291 QFEGEEEEDDEEDKEEEDEEEDDEDDDSNGEAEVDRKGK 407
Score = 37.7 bits (86), Expect = 0.006
Identities = 21/73 (28%), Positives = 35/73 (47%)
Frame = +3
Query: 351 IDEEDDEDEDEDEDEDEDEDDDDDEDEDEDEDEDEEKDSDTGSLELVDMKSETDQEADDE 410
+ DD E ++D + D+D + +E+ED+E+D K E D+E DDE
Sbjct: 216 VSATDDCSSQPKEAANDDNEPDNDVQFEGEEEEDDEED-----------KEEEDEEEDDE 362
Query: 411 VEINTGLGTSDRE 423
+ + G DR+
Sbjct: 363 DDDSNGEAEVDRK 401
Score = 36.2 bits (82), Expect = 0.018
Identities = 19/42 (45%), Positives = 27/42 (64%)
Frame = +3
Query: 339 DKDLISKVIEQIIDEEDDEDEDEDEDEDEDEDDDDDEDEDED 380
D D+ + E+ DEED E+EDE+ED DEDDD + + + D
Sbjct: 279 DNDVQFEGEEEEDDEEDKEEEDEEED---DEDDDSNGEAEVD 395
>TC14766 similar to UP|Q7XXR8 (Q7XXR8) Nascent polypeptide associated
complex alpha chain, partial (58%)
Length = 600
Score = 46.2 bits (108), Expect = 2e-05
Identities = 21/50 (42%), Positives = 36/50 (72%)
Frame = +2
Query: 340 KDLISKVIEQIIDEEDDEDEDEDEDEDEDEDDDDDEDEDEDEDEDEEKDS 389
++L++ +EQ ++ +DE ED+DED+DD+DD+DED+D E +E D+
Sbjct: 77 EELLAAHLEQ---QKIHDDEPVVEDDDEDDDDEDDDDEDDDNIEGQEGDA 217
Score = 44.3 bits (103), Expect = 7e-05
Identities = 18/38 (47%), Positives = 27/38 (70%)
Frame = +2
Query: 353 EEDDEDEDEDEDEDEDEDDDDDEDEDEDEDEDEEKDSD 390
E+ +DE ED+DEDDDD++D+DED+D E ++ D
Sbjct: 101 EQQKIHDDEPVVEDDDEDDDDEDDDDEDDDNIEGQEGD 214
Score = 42.7 bits (99), Expect = 2e-04
Identities = 17/52 (32%), Positives = 36/52 (68%)
Frame = +2
Query: 333 ITPEKVDKDLISKVIEQIIDEEDDEDEDEDEDEDEDEDDDDDEDEDEDEDED 384
+T + ++ L + + +Q I +++ ED+DED+D+++DDD+D+D E ++ D
Sbjct: 59 MTAQSQEELLAAHLEQQKIHDDEPVVEDDDEDDDDEDDDDEDDDNIEGQEGD 214
Score = 36.6 bits (83), Expect = 0.014
Identities = 15/49 (30%), Positives = 29/49 (58%)
Frame = +2
Query: 354 EDDEDEDEDEDEDEDEDDDDDEDEDEDEDEDEEKDSDTGSLELVDMKSE 402
E + D++ ++D++DDDDED+D+++D++ E S +SE
Sbjct: 101 EQQKIHDDEPVVEDDDEDDDDEDDDDEDDDNIEGQEGDASGRSKQTRSE 247
Score = 35.8 bits (81), Expect = 0.024
Identities = 14/45 (31%), Positives = 27/45 (59%)
Frame = +2
Query: 352 DEEDDEDEDEDEDEDEDEDDDDDEDEDEDEDEDEEKDSDTGSLEL 396
D+EDD+DED+D+++D++ + + + + EK S L+L
Sbjct: 143 DDEDDDDEDDDDEDDDNIEGQEGDASGRSKQTRSEKKSRKAMLKL 277
>BP043430
Length = 493
Score = 45.8 bits (107), Expect = 2e-05
Identities = 19/51 (37%), Positives = 35/51 (68%)
Frame = -1
Query: 352 DEEDDEDEDEDEDEDEDEDDDDDEDEDEDEDEDEEKDSDTGSLELVDMKSE 402
D+E +++E ++EDE+E++ +DDD+ + +E D+E+DSD L +K E
Sbjct: 349 DDEGNDNEVDEEDEEEEDSNDDDDGSESEEFNDDEEDSD*TQQILWSLKGE 197
Score = 37.7 bits (86), Expect = 0.006
Identities = 12/31 (38%), Positives = 25/31 (79%)
Frame = -1
Query: 351 IDEEDDEDEDEDEDEDEDEDDDDDEDEDEDE 381
+DEED+E+ED ++D+D E ++ ++DE++ +
Sbjct: 325 VDEEDEEEEDSNDDDDGSESEEFNDDEEDSD 233
Score = 37.4 bits (85), Expect = 0.008
Identities = 18/51 (35%), Positives = 32/51 (62%)
Frame = -1
Query: 360 DEDEDEDEDEDDDDDEDEDEDEDEDEEKDSDTGSLELVDMKSETDQEADDE 410
D++E +D DD+ +++E ++EDE+EE +D D SE+++ DDE
Sbjct: 379 DDNEGSGKDNDDEGNDNEVDEEDEEEEDSNDD------DDGSESEEFNDDE 245
Score = 36.6 bits (83), Expect = 0.014
Identities = 11/39 (28%), Positives = 28/39 (71%)
Frame = -1
Query: 352 DEEDDEDEDEDEDEDEDEDDDDDEDEDEDEDEDEEKDSD 390
D+ + +D D++ +++E D++DE+E++ D+D+ +S+
Sbjct: 379 DDNEGSGKDNDDEGNDNEVDEEDEEEEDSNDDDDGSESE 263
Score = 36.2 bits (82), Expect = 0.018
Identities = 15/51 (29%), Positives = 34/51 (66%)
Frame = -1
Query: 354 EDDEDEDEDEDEDEDEDDDDDEDEDEDEDEDEEKDSDTGSLELVDMKSETD 404
+D+E +D D++ ++++ D+EDE+E++ D++ S+ S E D + ++D
Sbjct: 379 DDNEGSGKDNDDEGNDNEVDEEDEEEEDSNDDDDGSE--SEEFNDDEEDSD 233
Score = 32.0 bits (71), Expect = 0.34
Identities = 12/33 (36%), Positives = 22/33 (66%)
Frame = -1
Query: 344 SKVIEQIIDEEDDEDEDEDEDEDEDEDDDDDED 376
++V E+ +EED D+D+ + +E DD++D D
Sbjct: 331 NEVDEEDEEEEDSNDDDDGSESEEFNDDEEDSD 233
Score = 30.8 bits (68), Expect = 0.76
Identities = 13/32 (40%), Positives = 23/32 (71%)
Frame = -1
Query: 347 IEQIIDEEDDEDEDEDEDEDEDEDDDDDEDED 378
+++ +EE+D ++D+D E E E +DD+ED D
Sbjct: 325 VDEEDEEEEDSNDDDDGSESE-EFNDDEEDSD 233
>TC11270 similar to UP|Q9LM88 (Q9LM88) F2D10.15, partial (9%)
Length = 735
Score = 45.4 bits (106), Expect = 3e-05
Identities = 19/36 (52%), Positives = 30/36 (82%)
Frame = +3
Query: 353 EEDDEDEDEDEDEDEDEDDDDDEDEDEDEDEDEEKD 388
EED+E+E+ +E+E+E+E +DE +EDEDEDEE++
Sbjct: 423 EEDEEEENGEEEEEEEEVVVEDEQGNEDEDEDEEEE 530
Score = 43.5 bits (101), Expect = 1e-04
Identities = 18/37 (48%), Positives = 30/37 (80%), Gaps = 3/37 (8%)
Frame = +3
Query: 353 EEDDEDEDEDEDEDEDED---DDDDEDEDEDEDEDEE 386
+E+DE+E+ E+E+E+E+ +D+ +EDEDEDE+EE
Sbjct: 420 QEEDEEEENGEEEEEEEEVVVEDEQGNEDEDEDEEEE 530
Score = 42.7 bits (99), Expect = 2e-04
Identities = 28/104 (26%), Positives = 55/104 (51%), Gaps = 18/104 (17%)
Frame = +3
Query: 337 KVDKDLISKVIEQIIDEEDDED---------------EDEDEDEDEDEDDDDDED---ED 378
K+ + L V E+ DEED+E+ ++EDE+E+ E+++++E+ ED
Sbjct: 309 KLVEKLNQGVEEEEEDEEDEEEQTPHPTRGRSRVEHAQEEDEEEENGEEEEEEEEVVVED 488
Query: 379 EDEDEDEEKDSDTGSLELVDMKSETDQEADDEVEINTGLGTSDR 422
E +EDE++D + + +V++K D +++G GT +
Sbjct: 489 EQGNEDEDEDEEEEGVVVVEVKG----RKVDSKGLHSGSGTPSK 608
Score = 37.0 bits (84), Expect = 0.011
Identities = 16/26 (61%), Positives = 20/26 (76%)
Frame = +2
Query: 351 IDEEDDEDEDEDEDEDEDEDDDDDED 376
ID D+EDEDEDEDED+DE + E+
Sbjct: 227 IDYLDEEDEDEDEDEDDDERGREKEE 304
Score = 35.4 bits (80), Expect = 0.031
Identities = 15/32 (46%), Positives = 22/32 (67%)
Frame = +2
Query: 361 EDEDEDEDEDDDDDEDEDEDEDEDEEKDSDTG 392
++EDEDEDED+DDDE E E+ +++ G
Sbjct: 239 DEEDEDEDEDEDDDERGREKEEAQAG*EAEPG 334
Score = 35.0 bits (79), Expect = 0.041
Identities = 16/34 (47%), Positives = 21/34 (61%)
Frame = +2
Query: 359 EDEDEDEDEDEDDDDDEDEDEDEDEDEEKDSDTG 392
++EDEDEDEDEDDD+ E E+ E + G
Sbjct: 239 DEEDEDEDEDEDDDERGREKEEAQAG*EAEPGGG 340
Score = 33.5 bits (75), Expect = 0.12
Identities = 13/22 (59%), Positives = 18/22 (81%)
Frame = +2
Query: 357 EDEDEDEDEDEDEDDDDDEDED 378
++EDEDEDEDED+D+ E E+
Sbjct: 239 DEEDEDEDEDEDDDERGREKEE 304
Score = 31.2 bits (69), Expect = 0.59
Identities = 12/26 (46%), Positives = 20/26 (76%)
Frame = +2
Query: 347 IEQIIDEEDDEDEDEDEDEDEDEDDD 372
I+ + +E++DEDEDED+DE E ++
Sbjct: 227 IDYLDEEDEDEDEDEDDDERGREKEE 304
>TC9022 similar to PIR|T46225|T46225 alpha NAC-like protein - Arabidopsis
thaliana {Arabidopsis thaliana;}, partial (75%)
Length = 1083
Score = 45.4 bits (106), Expect = 3e-05
Identities = 18/43 (41%), Positives = 30/43 (68%)
Frame = +3
Query: 353 EEDDEDEDEDEDEDEDEDDDDDEDEDEDEDEDEEKDSDTGSLE 395
+EDD ED +D+ ++ DDDEDED+D+++D+++D G E
Sbjct: 198 QEDDAPIVEDVKDDDKDEADDDEDEDDDDEDDDKEDGALGGSE 326
Score = 42.4 bits (98), Expect = 3e-04
Identities = 17/30 (56%), Positives = 26/30 (86%), Gaps = 1/30 (3%)
Frame = +3
Query: 350 IIDEEDDEDEDE-DEDEDEDEDDDDDEDED 378
I+++ D+D+DE D+DEDED+DD+DD+ ED
Sbjct: 216 IVEDVKDDDKDEADDDEDEDDDDEDDDKED 305
Score = 41.6 bits (96), Expect = 4e-04
Identities = 20/79 (25%), Positives = 42/79 (52%)
Frame = +3
Query: 318 LKDMIPGVKVKIFKVITPEKVDKDLISKVIEQIIDEEDDEDEDEDEDEDEDEDDDDDEDE 377
L+ ++ + + K P++ D ++ V + DE DD+++++D+DED+D++D
Sbjct: 144 LEQVLSAEETTLKKKPQPQEDDAPIVEDVKDDDKDEADDDEDEDDDDEDDDKEDGALGGS 323
Query: 378 DEDEDEDEEKDSDTGSLEL 396
+ + EK S L+L
Sbjct: 324 EGSKQSRSEKKSRKAMLKL 380
>BI420041
Length = 452
Score = 45.1 bits (105), Expect = 4e-05
Identities = 24/47 (51%), Positives = 34/47 (72%), Gaps = 6/47 (12%)
Frame = +2
Query: 346 VIEQIIDEEDDEDEDEDE---DEDEDED---DDDDEDEDEDEDEDEE 386
V E +DEE+D++ D +E DEDED D DDD+++E+E EDEDE+
Sbjct: 230 VQEVSMDEEEDDEFDYEEPLNDEDEDGDEYYDDDEQEEEEFEDEDED 370
Score = 43.1 bits (100), Expect = 1e-04
Identities = 19/50 (38%), Positives = 35/50 (70%), Gaps = 1/50 (2%)
Frame = +2
Query: 346 VIEQIIDEEDDEDEDEDEDEDEDEDDDDDEDEDEDEDEDEE-KDSDTGSL 394
+++++ +E+++DE + E+ DED+D DE D+DE E+EE +D D S+
Sbjct: 227 IVQEVSMDEEEDDEFDYEEPLNDEDEDGDEYYDDDEQEEEEFEDEDEDSV 376
Score = 42.7 bits (99), Expect = 2e-04
Identities = 23/54 (42%), Positives = 35/54 (64%), Gaps = 10/54 (18%)
Frame = +2
Query: 346 VIEQIIDEEDDEDEDEDED-------EDEDEDDD---DDEDEDEDEDEDEEKDS 389
V+E I +E DE+ED++ DEDED D DD++++E+E EDE++DS
Sbjct: 212 VLEPTIVQEVSMDEEEDDEFDYEEPLNDEDEDGDEYYDDDEQEEEEFEDEDEDS 373
Score = 28.5 bits (62), Expect = 3.8
Identities = 15/46 (32%), Positives = 26/46 (55%)
Frame = +2
Query: 367 EDEDDDDDEDEDEDEDEDEEKDSDTGSLELVDMKSETDQEADDEVE 412
++E+DD+ + E+ DEDE+ D E D + ++E +DE E
Sbjct: 248 DEEEDDEFDYEEPLNDEDEDGD------EYYDDDEQEEEEFEDEDE 367
>BP036655
Length = 556
Score = 44.7 bits (104), Expect = 5e-05
Identities = 25/78 (32%), Positives = 43/78 (55%)
Frame = -3
Query: 348 EQIIDEEDDEDEDEDEDEDEDEDDDDDEDEDEDEDEDEEKDSDTGSLELVDMKSETDQEA 407
E+ + ++ D+DE DE+E+ E+ DD+D ++DE +DE V+ +SET+ E
Sbjct: 407 EEGLSDDGDDDEVVDEEEEVVEEWSDDDDINDDESDDE-----------VEEESETEIET 261
Query: 408 DDEVEINTGLGTSDREDQ 425
+ E E G + ED+
Sbjct: 260 EKEEEEEEECGVENVEDE 207
Score = 42.0 bits (97), Expect = 3e-04
Identities = 19/50 (38%), Positives = 34/50 (68%), Gaps = 1/50 (2%)
Frame = -3
Query: 339 DKDLISKVIEQIIDE-EDDEDEDEDEDEDEDEDDDDDEDEDEDEDEDEEK 387
D D + E++++E DD+D ++DE +DE E++ + E E E E+E+EE+
Sbjct: 383 DDDEVVDEEEEVVEEWSDDDDINDDESDDEVEEESETEIETEKEEEEEEE 234
Score = 37.7 bits (86), Expect = 0.006
Identities = 21/60 (35%), Positives = 37/60 (61%), Gaps = 3/60 (5%)
Frame = -3
Query: 336 EKVDKDLISKVIEQIIDEED---DEDEDEDEDEDEDEDDDDDEDEDEDEDEDEEKDSDTG 392
E VD++ +V+E+ D++D DE +DE E+E E E + + E+E+E+E E + + G
Sbjct: 374 EVVDEE--EEVVEEWSDDDDINDDESDDEVEEESETEIETEKEEEEEEECGVENVEDEMG 201
Score = 35.8 bits (81), Expect = 0.024
Identities = 24/67 (35%), Positives = 32/67 (46%), Gaps = 2/67 (2%)
Frame = -3
Query: 363 EDEDEDEDDDDDEDEDEDEDEDEEKDSDTGSLELVDMKSETDQEADDEV--EINTGLGTS 420
E+E +D DDDE DE+E+ EE D D E+DDEV E T + T
Sbjct: 410 EEEGLSDDGDDDEVVDEEEEVVEEWSDDDDI---------NDDESDDEVEEESETEIETE 258
Query: 421 DREDQNE 427
E++ E
Sbjct: 257 KEEEEEE 237
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.314 0.135 0.390
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,394,378
Number of Sequences: 28460
Number of extensions: 147181
Number of successful extensions: 5787
Number of sequences better than 10.0: 241
Number of HSP's better than 10.0 without gapping: 1160
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3213
length of query: 700
length of database: 4,897,600
effective HSP length: 97
effective length of query: 603
effective length of database: 2,136,980
effective search space: 1288598940
effective search space used: 1288598940
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 58 (26.9 bits)
Lotus: description of TM0100a.10


