

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0090a.1
(150 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
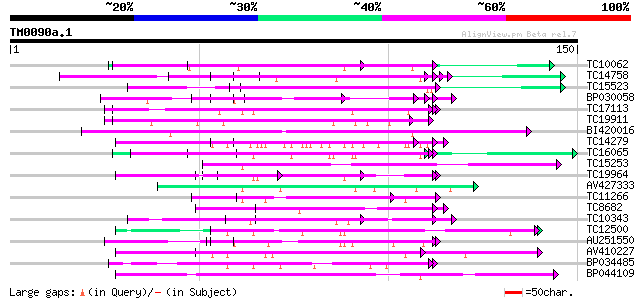
Score E
Sequences producing significant alignments: (bits) Value
TC10062 similar to BAD08700 (BAD08700) Cold shock domain protein... 67 2e-12
TC14758 similar to UP|O24601 (O24601) Glycine-rich RNA binding p... 62 5e-11
TC15523 similar to UP|GRPA_MAIZE (P10979) Glycine-rich RNA-bindi... 60 1e-10
BP030058 60 2e-10
TC17113 weakly similar to UP|Q9LFU8 (Q9LFU8) Proline-rich protei... 57 1e-09
TC19911 weakly similar to PIR|A44805|A44805 eggshell protein pre... 57 1e-09
BI420016 56 3e-09
TC14279 similar to UP|Q09085 (Q09085) Hydroxyproline-rich glycop... 55 5e-09
TC16065 weakly similar to UP|Q9SBM1 (Q9SBM1) Hydroxyproline-rich... 52 3e-08
TC15253 weakly similar to UP|Q9ZT16 (Q9ZT16) Arabinogalactan-pro... 50 1e-07
TC19964 similar to UP|Q40363 (Q40363) NuM1 protein, partial (11%) 50 2e-07
AV427333 49 3e-07
TC11266 weakly similar to UP|Q8L685 (Q8L685) Pherophorin-dz1 pro... 49 4e-07
TC8682 similar to UP|HPPD_SOLSC (Q9ARF9) 4-hydroxyphenylpyruvate... 49 5e-07
TC10343 similar to UP|Q9SZZ1 (Q9SZZ1) Fibrillarin-like protein (... 49 5e-07
TC12500 similar to GB|AAH45354.1|28279536|BC045354 FNBP4 protein... 48 6e-07
AU251550 48 8e-07
AV410227 47 1e-06
BP034485 47 1e-06
BP044109 47 2e-06
>TC10062 similar to BAD08700 (BAD08700) Cold shock domain protein 2, partial
(66%)
Length = 481
Score = 66.6 bits (161), Expect = 2e-12
Identities = 36/67 (53%), Positives = 37/67 (54%), Gaps = 1/67 (1%)
Frame = +3
Query: 48 QRGVENGGDDGGGGGGSGGGSGGDGVGSDGDDGDGCGSGGGGGGLGGDIGSGSDGDDG-G 106
+RG GG GGGG GGG GG G G G G G G GGG GG GG G G G G G
Sbjct: 273 RRGGGAGGGGRGGGGYGGGGYGGGGYGGGGRGGGGYGGGGGYGGSGGYGGGGGYGGGGRG 452
Query: 107 GGSGSGG 113
GG G GG
Sbjct: 453 GGRGGGG 473
Score = 63.9 bits (154), Expect = 1e-11
Identities = 46/128 (35%), Positives = 52/128 (39%), Gaps = 10/128 (7%)
Frame = +3
Query: 27 EGGGCRGGNSNAEWWLGRRR--RQRGVENGGDDGG-------GGGGSGGGSGGDGVGSDG 77
EG C G + E+ + R + +E G +G GGG GGG GG G G G
Sbjct: 153 EGFRCLGEGEDVEFIVASDPDGRAKAIEVTGPNGQAVQGTRRGGGAGGGGRGGGGYGGGG 332
Query: 78 DDGDGCGSGG-GGGGLGGDIGSGSDGDDGGGGSGSGGSTVEVMLMMVVAVRVVVAATVEG 136
G G G GG GGGG GG G G G GGGG GG G
Sbjct: 333 YGGGGYGGGGRGGGGYGGGGGYGGSGGYGGGGGYGGGG--------------------RG 452
Query: 137 GGCGGSDG 144
GG GG G
Sbjct: 453 GGRGGGGG 476
Score = 51.6 bits (122), Expect = 6e-08
Identities = 29/67 (43%), Positives = 30/67 (44%)
Frame = +3
Query: 28 GGGCRGGNSNAEWWLGRRRRQRGVENGGDDGGGGGGSGGGSGGDGVGSDGDDGDGCGSGG 87
GGG GG + G G GG GGG GG GG G G G G G G GG
Sbjct: 279 GGGAGGGGRGGGGYGGGGYGGGGYGGGGRGGGGYGGGGGYGGSGGYGGGGGYGGGGRGGG 458
Query: 88 GGGGLGG 94
GGG GG
Sbjct: 459 RGGGGGG 479
>TC14758 similar to UP|O24601 (O24601) Glycine-rich RNA binding protein 2,
partial (93%)
Length = 725
Score = 61.6 bits (148), Expect = 5e-11
Identities = 37/69 (53%), Positives = 37/69 (53%), Gaps = 9/69 (13%)
Frame = +1
Query: 54 GGDDGGGGGGSGGGSG------GDGVGSDGDDGDGCGSGGGGGGLGGDIGSGSDGDDGGG 107
GG G GGGG GGG G G G GS G G G G GGGGGG G G G DGGG
Sbjct: 331 GGGGGRGGGGYGGGGGRREGGYGGGGGSRGYGGGGGGYGGGGGGGYGGRREGGYGGDGGG 510
Query: 108 ---GSGSGG 113
GSG GG
Sbjct: 511 SRYGSGGGG 537
Score = 61.2 bits (147), Expect = 7e-11
Identities = 35/76 (46%), Positives = 38/76 (49%), Gaps = 3/76 (3%)
Frame = +1
Query: 43 GRRRRQRGVENGGDDGGGGGGSGGGSGGDGVGSDGDDGDGCGS---GGGGGGLGGDIGSG 99
GR + G GGGGG GGG GG G +G G G GS GGGGGG GG G G
Sbjct: 286 GRNITVNEAQARGGGGGGGGRGGGGYGGGGGRREGGYGGGGGSRGYGGGGGGYGGGGGGG 465
Query: 100 SDGDDGGGGSGSGGST 115
G GG G GG +
Sbjct: 466 YGGRREGGYGGDGGGS 513
Score = 57.0 bits (136), Expect = 1e-09
Identities = 39/98 (39%), Positives = 41/98 (41%)
Frame = +1
Query: 14 NIRFNSIMAMAVLEGGGCRGGNSNAEWWLGRRRRQRGVENGGDDGGGGGGSGGGSGGDGV 73
NI N A GGG RGG G RR+ G GG G GGG GG GG G
Sbjct: 292 NITVNEAQARGGGGGGGGRGGGGYGG---GGGRREGGYGGGGGSRGYGGGGGGYGGGGGG 462
Query: 74 GSDGDDGDGCGSGGGGGGLGGDIGSGSDGDDGGGGSGS 111
G G GG GGD G G GGGG G+
Sbjct: 463 GY---------GGRREGGYGGDGGGSRYGSGGGGGGGN 549
Score = 51.2 bits (121), Expect = 7e-08
Identities = 29/58 (50%), Positives = 30/58 (51%)
Frame = +1
Query: 60 GGGGSGGGSGGDGVGSDGDDGDGCGSGGGGGGLGGDIGSGSDGDDGGGGSGSGGSTVE 117
GGGG GGG GG G G G +G G GGGGG G G G GGGG G G E
Sbjct: 322 GGGGGGGGRGGGGYGGGGGRREG-GYGGGGGSRGY---GGGGGGYGGGGGGGYGGRRE 483
Score = 38.1 bits (87), Expect = 6e-04
Identities = 27/81 (33%), Positives = 31/81 (37%)
Frame = +1
Query: 67 GSGGDGVGSDGDDGDGCGSGGGGGGLGGDIGSGSDGDDGGGGSGSGGSTVEVMLMMVVAV 126
G+ DG ++ G GGGGGG GG G G GG G GGS
Sbjct: 271 GNELDGRNITVNEAQARGGGGGGGGRGGGGYGGGGGRREGGYGGGGGSR----------- 417
Query: 127 RVVVAATVEGGGCGGSDGRGH 147
GGG GG G G+
Sbjct: 418 ----GYGGGGGGYGGGGGGGY 468
>TC15523 similar to UP|GRPA_MAIZE (P10979) Glycine-rich RNA-binding,
abscisic acid-inducible protein, complete
Length = 972
Score = 60.5 bits (145), Expect = 1e-10
Identities = 31/56 (55%), Positives = 31/56 (55%)
Frame = +2
Query: 59 GGGGGSGGGSGGDGVGSDGDDGDGCGSGGGGGGLGGDIGSGSDGDDGGGGSGSGGS 114
GGGGG GG GG G G G SGGGGGG GG G G G GGG G GGS
Sbjct: 602 GGGGGGGGYGGGRREGGYGGGGGSRYSGGGGGGYGGSGGGGYGGRREGGGGGYGGS 769
Score = 53.1 bits (126), Expect = 2e-08
Identities = 36/85 (42%), Positives = 38/85 (44%), Gaps = 2/85 (2%)
Frame = +2
Query: 32 RGGNSNAEWWLGRRRRQRGVENGGDDGGGGGGSGGGSGGDGVGSDGDDGDGCGSGGGGGG 91
RGG + G RR G GGGGG G GG G G G G G GGGGG
Sbjct: 599 RGGGGGGGGYGGGRRE-------GGYGGGGGSRYSGGGGGGYGGSGGGGYGGRREGGGGG 757
Query: 92 LGGDIGSGSDGDDGGGGS--GSGGS 114
GG +GGGGS SGGS
Sbjct: 758 YGG-------SREGGGGSRYSSGGS 811
Score = 39.7 bits (91), Expect = 2e-04
Identities = 32/87 (36%), Positives = 35/87 (39%), Gaps = 1/87 (1%)
Frame = +2
Query: 62 GGSGGGSGGDGVGSDGDDGDGCGSGGGGGGLGGDIGSGSDGDDGGGGSG-SGGSTVEVML 120
G +G G V + G G GGGGGG GG G G GGGGS SGG
Sbjct: 536 GMNGQDIDGRNVTVNEAQARGRGGGGGGGGYGG--GRREGGYGGGGGSRYSGGG------ 691
Query: 121 MMVVAVRVVVAATVEGGGCGGSDGRGH 147
GGG GGS G G+
Sbjct: 692 ---------------GGGYGGSGGGGY 727
Score = 28.9 bits (63), Expect = 0.39
Identities = 20/46 (43%), Positives = 22/46 (47%), Gaps = 2/46 (4%)
Frame = +2
Query: 28 GGGCRGGNSNAEWWLGRRRRQRGVENGGDDGGGGGG--SGGGSGGD 71
GGG GG+ + G RR G GG GGGG S GGS D
Sbjct: 689 GGGGYGGSGGGGY--GGRREGGGGGYGGSREGGGGSRYSSGGSWRD 820
>BP030058
Length = 455
Score = 59.7 bits (143), Expect = 2e-10
Identities = 32/66 (48%), Positives = 37/66 (55%), Gaps = 1/66 (1%)
Frame = +1
Query: 25 VLEGGGCRGGN-SNAEWWLGRRRRQRGVENGGDDGGGGGGSGGGSGGDGVGSDGDDGDGC 83
+L GGG +G E + G GG+ GGGGGG GGG GGDG GS G G+G
Sbjct: 241 ILGGGGDKGLQFCGQEGYFG---------GGGEGGGGGGGDGGGGGGDGHGSGGGGGEGD 393
Query: 84 GSGGGG 89
G GGGG
Sbjct: 394 GGGGGG 411
Score = 54.7 bits (130), Expect = 7e-09
Identities = 31/62 (50%), Positives = 36/62 (58%), Gaps = 2/62 (3%)
Frame = +1
Query: 49 RGVENGGDDG--GGGGGSGGGSGGDGVGSDGDDGDGCGSGGGGGGLGGDIGSGSDGDDGG 106
+G++ G +G GGGG GGG GGDG G GDG GSGGGGG +GD GG
Sbjct: 262 KGLQFCGQEGYFGGGGEGGGGGGGDG---GGGGGDGHGSGGGGG----------EGDGGG 402
Query: 107 GG 108
GG
Sbjct: 403 GG 408
Score = 52.4 bits (124), Expect = 3e-08
Identities = 30/58 (51%), Positives = 30/58 (51%)
Frame = +1
Query: 54 GGDDGGGGGGSGGGSGGDGVGSDGDDGDGCGSGGGGGGLGGDIGSGSDGDDGGGGSGS 111
GGD G G G GG G G G GDG G GG G G GG G G DGGGG GS
Sbjct: 253 GGDKGLQFCGQEGYFGGGGEGGGGGGGDGGGGGGDGHGSGGGGGEG----DGGGGGGS 414
Score = 50.4 bits (119), Expect = 1e-07
Identities = 25/51 (49%), Positives = 26/51 (50%)
Frame = +1
Query: 63 GSGGGSGGDGVGSDGDDGDGCGSGGGGGGLGGDIGSGSDGDDGGGGSGSGG 113
G GG G G +G G G GGGGGG GG G G GGGG G GG
Sbjct: 247 GGGGDKGLQFCGQEGYFGGGGEGGGGGGGDGGGGGGDGHGSGGGGGEGDGG 399
Score = 50.1 bits (118), Expect = 2e-07
Identities = 27/59 (45%), Positives = 30/59 (50%)
Frame = +1
Query: 60 GGGGSGGGSGGDGVGSDGDDGDGCGSGGGGGGLGGDIGSGSDGDDGGGGSGSGGSTVEV 118
GGGG G G G G+G G GGG GG GG G GS G G G G GG + +V
Sbjct: 247 GGGGDKGLQFCGQEGYFGGGGEGGGGGGGDGGGGGGDGHGSGGGGGEGDGGGGGGSPQV 423
Score = 35.4 bits (80), Expect = 0.004
Identities = 21/49 (42%), Positives = 22/49 (44%)
Frame = +1
Query: 27 EGGGCRGGNSNAEWWLGRRRRQRGVENGGDDGGGGGGSGGGSGGDGVGS 75
EGGG GG+ G GGD G GGG G G GG G GS
Sbjct: 310 EGGGGGGGD--------------GGGGGGDGHGSGGGGGEGDGGGGGGS 414
>TC17113 weakly similar to UP|Q9LFU8 (Q9LFU8) Proline-rich protein, partial
(25%)
Length = 1077
Score = 57.0 bits (136), Expect = 1e-09
Identities = 41/108 (37%), Positives = 49/108 (44%), Gaps = 19/108 (17%)
Frame = -2
Query: 26 LEGGGCRGGNSNAEWWLGRRRRQRGVENGGDDGGGGGG-------SGGGSGGDGVGSDGD 78
+E GG GG +N +G +NGG GGGGGG G G+ G+G DG
Sbjct: 830 VEIGGLPGGGNNGG---------KGGKNGGGGGGGGGGVKPGITIGGKGNKGEGGCFDGG 678
Query: 79 DGDGCGSGGGG------------GGLGGDIGSGSDGDDGGGGSGSGGS 114
+G G GGG GGLGG G G G GG +GSG S
Sbjct: 677 GWNGLGINGGGLGGG*NGLGISGGGLGG*NGLGKSGGGLGGWNGSGKS 534
Score = 47.0 bits (110), Expect = 1e-06
Identities = 39/101 (38%), Positives = 44/101 (42%), Gaps = 16/101 (15%)
Frame = -2
Query: 28 GGGCRGGNSNAEWWLGRRRRQRGVENGGDDGGG-------GGGSGGGSGGDGVGSDGDDG 80
GGG GG + + + E G DGGG GGG GGG G G+ S G G
Sbjct: 773 GGGGGGGGGGVKPGITIGGKGNKGEGGCFDGGGWNGLGINGGGLGGG*NGLGI-SGGGLG 597
Query: 81 DGCGSGGGGGGLGGDIGSGSDGDD---------GGGGSGSG 112
G G GGGLGG GSG G GGG +G G
Sbjct: 596 G*NGLGKSGGGLGGWNGSGKSGGGLG*KGLGIRGGG*NGLG 474
Score = 46.6 bits (109), Expect = 2e-06
Identities = 40/96 (41%), Positives = 46/96 (47%), Gaps = 8/96 (8%)
Frame = -2
Query: 26 LEGGGCRGGNSNAEWWLGRRRRQRGVENG-GDDGGGGGG------SGGGSGGDGVGSDGD 78
+ GGG GG + LG G NG G GGG GG SGGG G G+G G
Sbjct: 659 INGGGLGGG*NG----LGISGGGLGG*NGLGKSGGGLGGWNGSGKSGGGLG*KGLGIRGG 492
Query: 79 DGDGCGSGGGG-GGLGGDIGSGSDGDDGGGGSGSGG 113
+G G GGG GLG + G G GGG +G GG
Sbjct: 491 G*NGLGINGGG*KGLGTNGG*NGLGIRGGGKNGIGG 384
Score = 30.8 bits (68), Expect = 0.10
Identities = 34/102 (33%), Positives = 40/102 (38%), Gaps = 11/102 (10%)
Frame = -2
Query: 26 LEGGGCRGGNSNAEWWLGRRRRQRGVENG-GDDGGG----------GGGSGGGSGGDGVG 74
+ GGG G N LG+ G NG G GGG GG +G G G G
Sbjct: 617 ISGGGLGG*NG-----LGKSGGGLGGWNGSGKSGGGLG*KGLGIRGGG*NGLGINGGG-- 459
Query: 75 SDGDDGDGCGSGGGGGGLGGDIGSGSDGDDGGGGSGSGGSTV 116
G G+ GG GL G G G +G G SG T+
Sbjct: 458 -----*KGLGTNGG*NGL-GIRGGGKNGIGGKKKKSSGSGTI 351
Score = 30.0 bits (66), Expect = 0.18
Identities = 17/52 (32%), Positives = 22/52 (41%)
Frame = -2
Query: 90 GGLGGDIGSGSDGDDGGGGSGSGGSTVEVMLMMVVAVRVVVAATVEGGGCGG 141
GGL G +G G GGG G GG V+ + + +GGG G
Sbjct: 821 GGLPGGGNNGGKGGKNGGGGGGGGGGVKPGITIGGKGNKGEGGCFDGGGWNG 666
Score = 28.5 bits (62), Expect = 0.51
Identities = 13/29 (44%), Positives = 14/29 (47%)
Frame = -2
Query: 88 GGGGLGGDIGSGSDGDDGGGGSGSGGSTV 116
GGG GG G G GGGG G T+
Sbjct: 809 GGGNNGGKGGKNGGGGGGGGGGVKPGITI 723
>TC19911 weakly similar to PIR|A44805|A44805 eggshell protein precursor -
fluke (Schistosoma haematobium) {Schistosoma
haematobium;} , partial (24%)
Length = 350
Score = 57.0 bits (136), Expect = 1e-09
Identities = 42/92 (45%), Positives = 49/92 (52%), Gaps = 7/92 (7%)
Frame = +1
Query: 28 GGGCRGGNS-NAEWWLGRRRRQ---RGVENGG-DDGGGGGGSGGGSGGDGVGSDGDDGDG 82
GGG G + +++ G RR + G+ GG DD GG GG GDG S G GD
Sbjct: 43 GGGSHGNDHRHSDNEHGGRRSETTGHGLSGGGYDDDNRSGGRTGGGYGDGGHSAGGYGDH 222
Query: 83 CG-SGGGGGGLG-GDIGSGSDGDDGGGGSGSG 112
G SGGGGGG G G + SGS G GG SG G
Sbjct: 223 GGRSGGGGGGYGDGGLSSGSGGYGDGGRSGVG 318
Score = 42.0 bits (97), Expect = 4e-05
Identities = 35/84 (41%), Positives = 37/84 (43%), Gaps = 2/84 (2%)
Frame = +1
Query: 26 LEGGGCRGGNSNAEWWLGRRRRQRGVENGGDDGGGGGGSGGGSGGDGVGSDGDDGDGCGS 85
L GGG N + R G +GG GG G GG SGG G G GDG S
Sbjct: 124 LSGGGYDDDNRSGG------RTGGGYGDGGHSAGGYGDHGGRSGGGG----GGYGDGGLS 273
Query: 86 GGGGG-GLGGDIGSG-SDGDDGGG 107
G GG G GG G G DG GG
Sbjct: 274 SGSGGYGDGGRSGVGYGDGXLSGG 345
Score = 35.4 bits (80), Expect = 0.004
Identities = 26/65 (40%), Positives = 31/65 (47%), Gaps = 7/65 (10%)
Frame = +1
Query: 56 DDGGGGGGSGGGS-GGDGVGSDGDDG----DGCGSGGGGGGLGGDIGSG--SDGDDGGGG 108
+ G G SGGGS G D SD + G + G G GGG D SG + G G GG
Sbjct: 16 NSAGNTGHSGGGSHGNDHRHSDNEHGGRRSETTGHGLSGGGYDDDNRSGGRTGGGYGDGG 195
Query: 109 SGSGG 113
+GG
Sbjct: 196 HSAGG 210
Score = 31.2 bits (69), Expect = 0.079
Identities = 24/75 (32%), Positives = 27/75 (36%), Gaps = 24/75 (32%)
Frame = +1
Query: 63 GSGGGSGGDGVGSD-----------------------GDDGDGCGSGGGGGGLG-GDIGS 98
G+ G SGG G+D G D D G GGG G G +
Sbjct: 25 GNTGHSGGGSHGNDHRHSDNEHGGRRSETTGHGLSGGGYDDDNRSGGRTGGGYGDGGHSA 204
Query: 99 GSDGDDGGGGSGSGG 113
G GD GG G GG
Sbjct: 205 GGYGDHGGRSGGGGG 249
>BI420016
Length = 559
Score = 55.8 bits (133), Expect = 3e-09
Identities = 41/125 (32%), Positives = 58/125 (45%), Gaps = 6/125 (4%)
Frame = -2
Query: 20 IMAMAVLEGGGCRGGNSNAEWW-----LGRRRRQRGVENGGDDGGGGGGSGGGSGGDGVG 74
++ +AV+E G G E +G R + VE + G G G GGG GG G G
Sbjct: 507 VVVVAVVEAGEVEAGEKREEGEDNEAEVGVRGEEGVVEPVAEGVGLGAGEGGGDGGVG-G 331
Query: 75 SDGDDGDGCGSGGGGGGLGGDIGSGSDGDDG-GGGSGSGGSTVEVMLMMVVAVRVVVAAT 133
G++G+ G G GG GG + G DG +G G+G G + +V +RV +
Sbjct: 330 EGGEEGESGAFGVGEGGEGGGVRVGEDGGEGVDAGTGFGDGFPP*VHWVVGFLRVWGSGG 151
Query: 134 VEGGG 138
GGG
Sbjct: 150 AHGGG 136
Score = 32.7 bits (73), Expect = 0.027
Identities = 22/52 (42%), Positives = 23/52 (43%), Gaps = 10/52 (19%)
Frame = -1
Query: 57 DGGGGGGSGGGSGGDGVGSDGDDGDGCGSGGGGG----------GLGGDIGS 98
D GG GGG GG G G + G GGGGG G GG IGS
Sbjct: 520 DHGGCCRRGGGGGGRGRGRREEGGRRGQRGGGGGQRRGRCRRASGRGGRIGS 365
Score = 29.6 bits (65), Expect = 0.23
Identities = 17/38 (44%), Positives = 17/38 (44%), Gaps = 3/38 (7%)
Frame = -1
Query: 79 DGDGC---GSGGGGGGLGGDIGSGSDGDDGGGGSGSGG 113
D GC G GGGG G G G G GGGG G
Sbjct: 520 DHGGCCRRGGGGGGRGRGRREEGGRRGQRGGGGGQRRG 407
>TC14279 similar to UP|Q09085 (Q09085) Hydroxyproline-rich glycoprotein
(HRGP) (Fragment), partial (57%)
Length = 941
Score = 55.1 bits (131), Expect = 5e-09
Identities = 45/118 (38%), Positives = 51/118 (43%), Gaps = 33/118 (27%)
Frame = -1
Query: 29 GGCRGGNSNAEWWLGRRRRQRGVE------NGGDDGGGGG------GSGGGSGGDGV--- 73
G GG +A W+ G + G GGDD GGGG G G G GG G+
Sbjct: 455 GDGEGGGGDA*WYGGGGDGEGGGGLT*W*GGGGDDVGGGGDL***GG*GMGDGGGGLL** 276
Query: 74 ---GSDGDDGDGC----GSGGGGGGLGGDI---GSGSDGDDGGG--------GSGSGG 113
G DGD G G G GG G G GGD+ G G + D GGG G G GG
Sbjct: 275 YGGGGDGDGGGGDL***GGGGDGDGGGGDL***GGGGECDGGGGDW**YGGGGEGDGG 102
Score = 46.2 bits (108), Expect = 2e-06
Identities = 44/121 (36%), Positives = 46/121 (37%), Gaps = 36/121 (29%)
Frame = -1
Query: 29 GGCRGGNSNAEWWLGRRRRQRGVENGGD---DGGGGGGSGG-------GSGGDGVGSD-- 76
G GG + W+ G G GGD G G G GG G GGDG G
Sbjct: 551 GEGEGGGGDV*WYGGGGNFTGG---GGDL*MYVGAGDGEGGGGDA*WYGGGGDGEGGGGL 381
Query: 77 ----GDDGDGCGSGG------------GGGGL-----GGDIGSGSDGD---DGGGGSGSG 112
G GD G GG GGGGL GG G G GD GGGG G G
Sbjct: 380 T*W*GGGGDDVGGGGDL***GG*GMGDGGGGLL**YGGGGDGDGGGGDL***GGGGDGDG 201
Query: 113 G 113
G
Sbjct: 200 G 198
Score = 43.5 bits (101), Expect = 2e-05
Identities = 31/65 (47%), Positives = 32/65 (48%), Gaps = 10/65 (15%)
Frame = -1
Query: 54 GGDDGGGGGGS---GGGSGGDGVGSDG--DDGDGCGSGGGG-----GGLGGDIGSGSDGD 103
GGD GGGG GGG DG G D G G G GGGG GG GGD+ G
Sbjct: 218 GGDGDGGGGDL***GGGGECDGGGGDW**YGGGGEGDGGGGDL**YGGGGGDL**YGFGT 39
Query: 104 DGGGG 108
GGGG
Sbjct: 38 GGGGG 24
Score = 42.7 bits (99), Expect = 3e-05
Identities = 35/83 (42%), Positives = 37/83 (44%), Gaps = 29/83 (34%)
Frame = -1
Query: 60 GGGGSG---GGSGGDGVGSDGD------DGDGCGSGG-----GGGGL----GGDI----- 96
GGGG G GGD VG GD G+G G GG GGGG GGD+
Sbjct: 638 GGGGEA*M*TGGGGDFVGGGGDL*IYVGAGEGEGGGGDV*WYGGGGNFTGGGGDL*MYVG 459
Query: 97 ---GSGSDGDD---GGGGSGSGG 113
G G GD GGGG G GG
Sbjct: 458 AGDGEGGGGDA*WYGGGGDGEGG 390
Score = 40.0 bits (92), Expect = 2e-04
Identities = 35/87 (40%), Positives = 35/87 (40%), Gaps = 24/87 (27%)
Frame = -1
Query: 54 GGDDGGGGG--------GSGGGSGGD------GVGSDGDDGD-----GCGSGGGGGGLGG 94
GGD GGGG G G G GGD G G GD G G G GGGG
Sbjct: 602 GGDFVGGGGDL*IYVGAGEGEGGGGDV*WYGGGGNFTGGGGDL*MYVGAGDGEGGGGDA* 423
Query: 95 DIGSGSDGDDGGGGS-----GSGGSTV 116
G G GD GGG G GG V
Sbjct: 422 WYGGG--GDGEGGGGLT*W*GGGGDDV 348
Score = 36.2 bits (82), Expect = 0.002
Identities = 24/48 (50%), Positives = 26/48 (54%), Gaps = 8/48 (16%)
Frame = -1
Query: 52 ENGGDD----GGGGGGSGGGSGGDGVGSDGDDGD----GCGSGGGGGG 91
+ GG D GGGG G GG GGD G GD G G+GGGGGG
Sbjct: 158 DGGGGDW**YGGGGEGDGG--GGDL**YGGGGGDL**YGFGTGGGGGG 21
Score = 30.0 bits (66), Expect = 0.18
Identities = 22/54 (40%), Positives = 24/54 (43%), Gaps = 10/54 (18%)
Frame = -1
Query: 28 GGGCRGGNSNAEWWLGRRRRQRGVENGGD---DGGGGG-------GSGGGSGGD 71
GG C GG + W G GGD GGGGG G+GGG GGD
Sbjct: 170 GGECDGGGGD---W**YGGGGEGDGGGGDL**YGGGGGDL**YGFGTGGGGGGD 18
>TC16065 weakly similar to UP|Q9SBM1 (Q9SBM1) Hydroxyproline-rich
glycoprotein DZ-HRGP precursor, partial (20%)
Length = 608
Score = 52.4 bits (124), Expect = 3e-08
Identities = 38/91 (41%), Positives = 40/91 (43%), Gaps = 11/91 (12%)
Frame = -1
Query: 33 GGNSNAEWWLGRRRRQRGVENGGDDGGGGGG-SGGGSGGDGV------GSDGDDGDGCG- 84
GG + G +G GG GGGG G GGG GG GV G G G G
Sbjct: 368 GGGPGPQGGRGYGGPPQGGRGGGYGGGGGPGYGGGGRGGHGVPQQQYGGPPEYQGRGRGG 189
Query: 85 ---SGGGGGGLGGDIGSGSDGDDGGGGSGSG 112
GG GG GG G G G GGGG GSG
Sbjct: 188 PSQQGGRGGYSGGGGGYGGGGGRGGGGMGSG 96
Score = 50.1 bits (118), Expect = 2e-07
Identities = 35/82 (42%), Positives = 35/82 (42%), Gaps = 16/82 (19%)
Frame = -1
Query: 48 QRGVENGGDDGGGGGGSGGGSGGDGVGSDGDDGDGC------------GSGGGG----GG 91
Q G GG GG GG GG GG G G G G G G G GG GG
Sbjct: 350 QGGRGYGGPPQGGRGGGYGGGGGPGYGGGGRGGHGVPQQQYGGPPEYQGRGRGGPSQQGG 171
Query: 92 LGGDIGSGSDGDDGGGGSGSGG 113
GG G G G GGGG G GG
Sbjct: 170 RGGYSGGGG-GYGGGGGRGGGG 108
Score = 43.9 bits (102), Expect = 1e-05
Identities = 36/98 (36%), Positives = 39/98 (39%), Gaps = 8/98 (8%)
Frame = -1
Query: 61 GGGSGGGSGGDGVGSDGDDGDGCGSGGGGG---GLGGDIGSGSDGDDGGG-----GSGSG 112
GGG G GG G G G G G GGGGG G GG G G GG G G G
Sbjct: 371 GGGGPGPQGGRGYGGPPQGGRGGGYGGGGGPGYGGGGRGGHGVPQQQYGGPPEYQGRGRG 192
Query: 113 GSTVEVMLMMVVAVRVVVAATVEGGGCGGSDGRGHDSM 150
G + + + GGG GG GRG M
Sbjct: 191 GPSQQ---------GGRGGYSGGGGGYGGGGGRGGGGM 105
Score = 43.9 bits (102), Expect = 1e-05
Identities = 30/84 (35%), Positives = 34/84 (39%)
Frame = -1
Query: 28 GGGCRGGNSNAEWWLGRRRRQRGVENGGDDGGGGGGSGGGSGGDGVGSDGDDGDGCGSGG 87
GGG RGG+ + G +G GG GG G G GG G GG
Sbjct: 272 GGGGRGGHGVPQQQYGGPPEYQGRGRGGPSQQGGRGGYSGGGG-------------GYGG 132
Query: 88 GGGGLGGDIGSGSDGDDGGGGSGS 111
GGG GG +GSG GG S
Sbjct: 131 GGGRGGGGMGSGRGVSPSHGGPPS 60
>TC15253 weakly similar to UP|Q9ZT16 (Q9ZT16) Arabinogalactan-protein
(ATAGP4) (AT5G10430/F12B17_220), partial (42%)
Length = 575
Score = 50.4 bits (119), Expect = 1e-07
Identities = 32/104 (30%), Positives = 47/104 (44%), Gaps = 9/104 (8%)
Frame = -2
Query: 52 ENGGDDGGG---------GGGSGGGSGGDGVGSDGDDGDGCGSGGGGGGLGGDIGSGSDG 102
EN + GGG G G +G DG+G+ G++ +G G+ +G +G G+ G
Sbjct: 466 ENAAEGGGGVSALFGPGPAAGPGPDAGPDGIGAGGEEAEGVGA-----PVGEAVGVGAGG 302
Query: 103 DDGGGGSGSGGSTVEVMLMMVVAVRVVVAATVEGGGCGGSDGRG 146
+ G G +GG +VV V V GGG G + G G
Sbjct: 301 EAEGAGVAAGG--------VVVGAGVAAGGGVAGGGAGAAAGGG 194
Score = 35.0 bits (79), Expect = 0.005
Identities = 23/72 (31%), Positives = 31/72 (42%), Gaps = 5/72 (6%)
Frame = -2
Query: 50 GVENGGDDGGGGGGSGG-----GSGGDGVGSDGDDGDGCGSGGGGGGLGGDIGSGSDGDD 104
G+ GG++ G G G G+GG+ +G G +GG G G G G G
Sbjct: 379 GIGAGGEEAEGVGAPVGEAVGVGAGGEA------EGAGVAAGGVVVGAGVAAGGGVAGGG 218
Query: 105 GGGGSGSGGSTV 116
G +G GG V
Sbjct: 217 AGAAAGGGGGVV 182
Score = 33.5 bits (75), Expect = 0.016
Identities = 21/50 (42%), Positives = 27/50 (54%), Gaps = 1/50 (2%)
Frame = -2
Query: 50 GVENGGDDGGGGGGSGGGSGGDGVGSDGD-DGDGCGSGGGGGGLGGDIGS 98
GV GG+ G G +GG G GV + G G G G+ GGGG G +G+
Sbjct: 319 GVGAGGEAEGAGVAAGGVVVGAGVAAGGGVAGGGAGAAAGGGG-GVVVGA 173
Score = 29.6 bits (65), Expect = 0.23
Identities = 20/45 (44%), Positives = 22/45 (48%), Gaps = 3/45 (6%)
Frame = -1
Query: 50 GVENGGDDGGGGGG-SGGGSGGDGV--GSDGDDGDGCGSGGGGGG 91
G + GG G G GG +GGGSG G G G G S GG G
Sbjct: 383 GWDWGGWRGSGRGGCTGGGSGWSGSWRGGGGSRGGSWWSSGGSRG 249
Score = 27.7 bits (60), Expect = 0.88
Identities = 19/46 (41%), Positives = 19/46 (41%), Gaps = 3/46 (6%)
Frame = -1
Query: 38 AEW-WLGRRRRQRGVENGGDDGGGGG--GSGGGSGGDGVGSDGDDG 80
A W W G R RG GG G G G GG GG S G G
Sbjct: 386 AGWDWGGWRGSGRGGCTGGGSGWSGSWRGGGGSRGGSWWSSGGSRG 249
>TC19964 similar to UP|Q40363 (Q40363) NuM1 protein, partial (11%)
Length = 562
Score = 50.1 bits (118), Expect = 2e-07
Identities = 30/59 (50%), Positives = 31/59 (51%), Gaps = 1/59 (1%)
Frame = +2
Query: 56 DDGGGGGGSGGGSGGDGVGSDGDDGDGCGSGG-GGGGLGGDIGSGSDGDDGGGGSGSGG 113
D+ G GG SGGG G G G D G G G G G GG GGD G G GGSG GG
Sbjct: 128 DNQGSGGRSGGGRSGGGRGGRFDSGRGGGRGRFGSGGRGGDRG-------GRGGSGGGG 283
Score = 45.4 bits (106), Expect = 4e-06
Identities = 31/65 (47%), Positives = 33/65 (50%), Gaps = 2/65 (3%)
Frame = +2
Query: 52 ENGGDDGGGGGG-SGGGSGGDGVGSDGDDGDGCGSGG-GGGGLGGDIGSGSDGDDGGGGS 109
+N G G GGG SGGG GG D G G G G G GG GGD G G GGGG
Sbjct: 128 DNQGSGGRSGGGRSGGGRGG-----RFDSGRGGGRGRFGSGGRGGD--RGGRGGSGGGGR 286
Query: 110 GSGGS 114
G G+
Sbjct: 287 GGRGT 301
Score = 41.2 bits (95), Expect = 8e-05
Identities = 25/50 (50%), Positives = 25/50 (50%), Gaps = 5/50 (10%)
Frame = +2
Query: 50 GVENGGDDGGGGGGS-----GGGSGGDGVGSDGDDGDGCGSGGGGGGLGG 94
G GG GGG GG GGG G G G G D G G G GGGG GG
Sbjct: 146 GRSGGGRSGGGRGGRFDSGRGGGRGRFGSGGRGGDRGGRG-GSGGGGRGG 292
Score = 37.7 bits (86), Expect = 8e-04
Identities = 22/44 (50%), Positives = 24/44 (54%)
Frame = +2
Query: 29 GGCRGGNSNAEWWLGRRRRQRGVENGGDDGGGGGGSGGGSGGDG 72
GG RGG ++ GR R G GGD GG GG GGG GG G
Sbjct: 170 GGGRGGRFDSGRGGGRGRFGSG-GRGGDRGGRGGSGGGGRGGRG 298
Score = 31.6 bits (70), Expect = 0.061
Identities = 17/45 (37%), Positives = 20/45 (43%)
Frame = +2
Query: 28 GGGCRGGNSNAEWWLGRRRRQRGVENGGDDGGGGGGSGGGSGGDG 72
GGG GG + GR + +GG G GG G G GG G
Sbjct: 155 GGGRSGGGRGGRFDSGRGGGRGRFGSGGRGGDRGGRGGSGGGGRG 289
>AV427333
Length = 387
Score = 49.3 bits (116), Expect = 3e-07
Identities = 40/118 (33%), Positives = 48/118 (39%), Gaps = 33/118 (27%)
Frame = -3
Query: 40 WWLGRRRRQRGVENGGDDGGG-------GGGSGGGSGG-----DGVGSDGDDGDGCGSGG 87
WW G G G GGG GGG G GG DG G D G+GC GG
Sbjct: 379 WWFGEGEGGSGGGGGRTCGGGVIG*CGDGGGRTCGGGGVNG*DDGGGGDDGGGEGCPRGG 200
Query: 88 GGG---GLGGD------IGSGSDGDDGG-----GGSGSGGST-------VEVMLMMVV 124
G G GGD +G G G +GG G G GG VE +L++++
Sbjct: 199 GANE*EGGGGDLK*WEGVGGGLKGWEGGVGGL*GWEGGGGKCGK*KLG*VEAILLLLL 26
>TC11266 weakly similar to UP|Q8L685 (Q8L685) Pherophorin-dz1 protein
precursor, partial (6%)
Length = 354
Score = 48.9 bits (115), Expect = 4e-07
Identities = 29/67 (43%), Positives = 34/67 (50%), Gaps = 13/67 (19%)
Frame = -2
Query: 49 RGVENGGDDGGG----------GGGSGG---GSGGDGVGSDGDDGDGCGSGGGGGGLGGD 95
+G E GG+ GGG GGG GG G+GG+G DG DG G G GG+GG
Sbjct: 203 KGEEAGGEPGGGNKGRKGKGETGGGEGGVKPGTGGNGKNGDGPDGGG*SGLGMSGGVGG* 24
Query: 96 IGSGSDG 102
G G G
Sbjct: 23 RGLGMSG 3
Score = 38.5 bits (88), Expect = 5e-04
Identities = 26/56 (46%), Positives = 30/56 (53%), Gaps = 2/56 (3%)
Frame = -2
Query: 61 GGGSGGGSGGDGVGSDGDDGDGCGSGGGGGGLGGDIGSGSDGD--DGGGGSGSGGS 114
G +GG GG G+ G G G GG GG G G+G +GD DGGG SG G S
Sbjct: 200 GEEAGGEPGG---GNKGRKGKGETGGGEGGVKPGTGGNGKNGDGPDGGG*SGLGMS 42
>TC8682 similar to UP|HPPD_SOLSC (Q9ARF9) 4-hydroxyphenylpyruvate
dioxygenase (4HPPD) (HPD) (HPPDase) , partial (29%)
Length = 576
Score = 48.5 bits (114), Expect = 5e-07
Identities = 27/49 (55%), Positives = 27/49 (55%), Gaps = 1/49 (2%)
Frame = -2
Query: 66 GGSGGDGVGS-DGDDGDGCGSGGGGGGLGGDIGSGSDGDDGGGGSGSGG 113
GG G DGVG DGD DG GGG G GG G GD GGGG G G
Sbjct: 563 GGEGLDGVGDFDGDGADGEAVGGGEGEAGG---GGESGDGGGGGEGDFG 426
Score = 38.9 bits (89), Expect = 4e-04
Identities = 26/67 (38%), Positives = 31/67 (45%)
Frame = -2
Query: 50 GVENGGDDGGGGGGSGGGSGGDGVGSDGDDGDGCGSGGGGGGLGGDIGSGSDGDDGGGGS 109
GV + DG G GGG G G G G+ GDG GGGG G G + G + + G
Sbjct: 545 GVGDFDGDGADGEAVGGGEGEAGGG--GESGDG---GGGGEGDFGRVWGGEEEAEVAGAE 381
Query: 110 GSGGSTV 116
GG V
Sbjct: 380 EVGGVRV 360
Score = 36.2 bits (82), Expect = 0.002
Identities = 22/51 (43%), Positives = 25/51 (48%), Gaps = 3/51 (5%)
Frame = -2
Query: 60 GGGGSGGGSGGDGVGSDGD---DGDGCGSGGGGGGLGGDIGSGSDGDDGGG 107
GG G G DG G+DG+ G+G GGG G GG G G G GG
Sbjct: 563 GGEGLDGVGDFDGDGADGEAVGGGEGEAGGGGESGDGGGGGEGDFGRVWGG 411
>TC10343 similar to UP|Q9SZZ1 (Q9SZZ1) Fibrillarin-like protein (Fibrillarin
2) (AtFib2), partial (85%)
Length = 1116
Score = 48.5 bits (114), Expect = 5e-07
Identities = 32/66 (48%), Positives = 33/66 (49%), Gaps = 5/66 (7%)
Frame = +1
Query: 58 GGGGGGSGGGSGGDGVGSDGDDGDGCGS-----GGGGGGLGGDIGSGSDGDDGGGGSGSG 112
GGGGG G G G G G G G G G+ GGG GG GG G G G G GG G
Sbjct: 55 GGGGGFRGRGDRGRGRGDGGGRGGGRGTPFKARGGGRGGGGGRGGRGG-GRGGRGGGMKG 231
Query: 113 GSTVEV 118
GS V V
Sbjct: 232 GSRVVV 249
Score = 43.9 bits (102), Expect = 1e-05
Identities = 30/67 (44%), Positives = 33/67 (48%), Gaps = 4/67 (5%)
Frame = +1
Query: 32 RGGNSNAEWWLGRRRRQRGVENGGDDGGGGG----GSGGGSGGDGVGSDGDDGDGCGSGG 87
RGG + GR R RG +GG GGG G GGG GG G G G G G GG
Sbjct: 52 RGGGGG---FRGRGDRGRGRGDGGGRGGGRGTPFKARGGGRGGGG----GRGGRGGGRGG 210
Query: 88 GGGGLGG 94
GGG+ G
Sbjct: 211 RGGGMKG 231
Score = 41.6 bits (96), Expect = 6e-05
Identities = 23/49 (46%), Positives = 24/49 (48%), Gaps = 1/49 (2%)
Frame = +1
Query: 66 GGSGGDGVGSDGDDGDGCGSGGG-GGGLGGDIGSGSDGDDGGGGSGSGG 113
G GG G GD G G G GGG GGG G + G GGGG G G
Sbjct: 49 GRGGGGGFRGRGDRGRGRGDGGGRGGGRGTPFKARGGGRGGGGGRGGRG 195
Score = 35.4 bits (80), Expect = 0.004
Identities = 26/64 (40%), Positives = 26/64 (40%), Gaps = 1/64 (1%)
Frame = +1
Query: 84 GSGGGGGGLG-GDIGSGSDGDDGGGGSGSGGSTVEVMLMMVVAVRVVVAATVEGGGCGGS 142
G GGGGG G GD G G GD GG G G G GGG GG
Sbjct: 49 GRGGGGGFRGRGDRGRGR-GDGGGRGGGRG-----------------TPFKARGGGRGGG 174
Query: 143 DGRG 146
GRG
Sbjct: 175 GGRG 186
Score = 28.5 bits (62), Expect = 0.51
Identities = 23/62 (37%), Positives = 26/62 (41%), Gaps = 1/62 (1%)
Frame = +1
Query: 29 GGCRGGNSNAEWWLGRRRRQRGVENGGDDGGGGGGSGGG-SGGDGVGSDGDDGDGCGSGG 87
GG RGG + R G GG GGG GG GGG GG V + +G
Sbjct: 109 GGGRGGGRGTPFKARGGGRGGGGGRGGR-GGGRGGRGGGMKGGSRVVVEPHRHEGIFIAK 285
Query: 88 GG 89
GG
Sbjct: 286 GG 291
>TC12500 similar to GB|AAH45354.1|28279536|BC045354 FNBP4 protein {Danio
rerio;} , partial (5%)
Length = 678
Score = 48.1 bits (113), Expect = 6e-07
Identities = 40/98 (40%), Positives = 46/98 (46%), Gaps = 11/98 (11%)
Frame = -2
Query: 54 GGDDGGGGGGSGGGSGGDGVGSDGDDGDGCGSG---------GGGGGLGGDIGSGSDGDD 104
GG GGGGGG G GG G SD D GSG GGGG + G DG
Sbjct: 674 GGVGGGGGGGDRVGVGG-GASSDVSGVDSRGSGVLSIEDIGRGGGGMVATPYLGGGDGGC 498
Query: 105 GGGGSGSGGSTVEVMLMMVVAVRVVVA--ATVEGGGCG 140
GG GSG V V+++ + VV+ VEGGG G
Sbjct: 497 GGEGSG-----VWVIVVRELGGDGVVSDVRCVEGGGSG 399
Score = 47.0 bits (110), Expect = 1e-06
Identities = 47/149 (31%), Positives = 57/149 (37%), Gaps = 36/149 (24%)
Frame = -2
Query: 29 GGCRGGNSNAEWWLGRRRRQRGVENGGD---------DGGGGGG------SGGGSGGDGV 73
GGC GG + W + R E GGD +GGG G S GG GG V
Sbjct: 506 GGC-GGEGSGVWVIVVR------ELGGDGVVSDVRCVEGGGSGALSVADMSKGGEGGMVV 348
Query: 74 GSD-GDDGDGCGSGG--------------------GGGGLGGDIGSGSDGDDGGGGSGSG 112
G G GCG G GGG +GG G G G G G G
Sbjct: 347 TPYFGGGGGGCGGDGREQGKEASPDIIALPAITEAGGGVVGGRDGGGGCGGGGEGEQGRE 168
Query: 113 GSTVEVMLMMVVAVRVVVAATVEGGGCGG 141
S ++ ++A+ + A EGGG GG
Sbjct: 167 TSPTDLETFEIIALPAIKGA--EGGGGGG 87
Score = 37.4 bits (85), Expect = 0.001
Identities = 28/74 (37%), Positives = 33/74 (43%), Gaps = 9/74 (12%)
Frame = -2
Query: 54 GGDDGGGGGGSGGGSGGDGVGSDGDDGDGC---------GSGGGGGGLGGDIGSGSDGDD 104
GG DGGGG G GGG G G + D + G+ GGGGG GG S G +
Sbjct: 227 GGRDGGGGCG-GGGEGEQGRETSPTDLETFEIIALPAIKGAEGGGGG-GGRALSDFGGVE 54
Query: 105 GGGGSGSGGSTVEV 118
GGG + V
Sbjct: 53 GGGSGALSAEDIGV 12
Score = 31.2 bits (69), Expect = 0.079
Identities = 27/80 (33%), Positives = 30/80 (36%), Gaps = 10/80 (12%)
Frame = -2
Query: 28 GGGCRGGNSNAEWWLGRRRRQRGVEN---------GGDDGGGGGGSGGGSGGDGVGSDGD 78
GGGC GG + GR +E G +GGGGGG S GV
Sbjct: 212 GGGCGGGGEGEQ---GRETSPTDLETFEIIALPAIKGAEGGGGGGGRALSDFGGV----- 57
Query: 79 DGDGCGSGGGGGGLGG-DIG 97
GGG G L DIG
Sbjct: 56 ------EGGGSGALSAEDIG 15
>AU251550
Length = 329
Score = 47.8 bits (112), Expect = 8e-07
Identities = 32/68 (47%), Positives = 33/68 (48%), Gaps = 8/68 (11%)
Frame = -3
Query: 54 GGDDG-GGGGGSGGGSGGDGVGSDGDDGDGCGSGG---GGGGLGGDIGSGSDGDDGGGG- 108
GG G GG GG GGG G + G DG CG GG GG GG GS G GGGG
Sbjct: 309 GGCGGCGGPGGPGGGGG**NMAGGGGDGGPCGGGGW*CPNGGGGGAGA*GSRGGPGGGGL 130
Query: 109 ---SGSGG 113
G GG
Sbjct: 129 **PGGCGG 106
Score = 45.1 bits (105), Expect = 5e-06
Identities = 30/65 (46%), Positives = 32/65 (49%), Gaps = 5/65 (7%)
Frame = -3
Query: 54 GGDDGGGGGGSGGGSGGDGVGSDGDDGDGCGSGGGG-----GGLGGDIGSGSDGDDGGGG 108
GG GGGG + G GGDG G G G C +GGGG G GG G G G GG
Sbjct: 282 GGPGGGGG**NMAGGGGDG-GPCGGGGW*CPNGGGGGAGA*GSRGGPGGGGL**PGGCGG 106
Query: 109 SGSGG 113
G GG
Sbjct: 105 PGGGG 91
Score = 44.3 bits (103), Expect = 9e-06
Identities = 31/71 (43%), Positives = 32/71 (44%), Gaps = 10/71 (14%)
Frame = -3
Query: 53 NGGDDGGGGGGSGGG------SGGDGVGSDGDDGDGCGSGGGG----GGLGGDIGSGSDG 102
N GG GG GGG GG G G+ G G G GGGG GG GG G G
Sbjct: 252 NMAGGGGDGGPCGGGGW*CPNGGGGGAGA*GSRG---GPGGGGL**PGGCGGPGGGGLW* 82
Query: 103 DDGGGGSGSGG 113
GGG G GG
Sbjct: 81 PGGGGPGGPGG 49
Score = 42.0 bits (97), Expect = 4e-05
Identities = 36/88 (40%), Positives = 38/88 (42%)
Frame = -3
Query: 26 LEGGGCRGGNSNAEWWLGRRRRQRGVENGGDDGGGGGGSGGGSGGDGVGSDGDDGDGCGS 85
+ GGG GG W NGG GGG G+ G GG G G G GCG
Sbjct: 249 MAGGGGDGGPCGGGGW*--------CPNGG---GGGAGA*GSRGGPGGGGL**PG-GCG- 109
Query: 86 GGGGGGLGGDIGSGSDGDDGGGGSGSGG 113
G GGGGL G G G GG G GG
Sbjct: 108 GPGGGGLW---*PGGGGPGGPGGPGCGG 34
Score = 41.6 bits (96), Expect = 6e-05
Identities = 28/61 (45%), Positives = 31/61 (49%), Gaps = 6/61 (9%)
Frame = -3
Query: 60 GGGGSGGGSGGDGVGSDGDDGDGCGSG----GGGGGLGGDIGSGS--DGDDGGGGSGSGG 113
GGG GG GG G G G G G G GGGG GG G G + GGGG+G+ G
Sbjct: 327 GGGPGPGGCGGCG----GPGGPGGGGG**NMAGGGGDGGPCGGGGW*CPNGGGGGAGA*G 160
Query: 114 S 114
S
Sbjct: 159 S 157
>AV410227
Length = 428
Score = 47.4 bits (111), Expect = 1e-06
Identities = 35/96 (36%), Positives = 45/96 (46%), Gaps = 14/96 (14%)
Frame = -1
Query: 29 GGCRGGNSNAEWWLGRRRRQRGVE------NGGD---DGGGGGGSGG---GSGGDGVGSD 76
GG RGG+ R G E GGD +GGG G+GG G GG+ G
Sbjct: 428 GGKRGGDGEFAHGGAERGSTAGGEPAYEGGGGGD*AEEGGGVDGAGGVMEGGGGEEAGDG 249
Query: 77 GDDGDGCGSGGGGGGLGGDIGSGSDGD--DGGGGSG 110
G++G G G G G GD+G ++ + DGGG G
Sbjct: 248 GEEGGGVGVGVVGEVERGDVGDVAEEEAVDGGGRGG 141
Score = 45.1 bits (105), Expect = 5e-06
Identities = 34/96 (35%), Positives = 42/96 (43%), Gaps = 4/96 (4%)
Frame = -1
Query: 50 GVENGGDDGGGGGGSGGGS--GGDGVGSDGDDGDGCGSGGGGGGLGGDIGSGSDGDDGGG 107
G + GGD GG+ GS GG+ G GD GGG G GG + G + G G
Sbjct: 428 GGKRGGDGEFAHGGAERGSTAGGEPAYEGGGGGD*AEEGGGVDGAGGVMEGGGGEEAGDG 249
Query: 108 GSGSGGSTVEVM--LMMVVAVRVVVAATVEGGGCGG 141
G GG V V+ + V V+GGG GG
Sbjct: 248 GEEGGGVGVGVVGEVERGDVGDVAEEEAVDGGGRGG 141
>BP034485
Length = 485
Score = 47.4 bits (111), Expect = 1e-06
Identities = 32/96 (33%), Positives = 41/96 (42%), Gaps = 9/96 (9%)
Frame = +3
Query: 27 EGGGCRGGNSNAEWWLGRRRRQRGVENGGDDGGGGGGSGGGSGG---DGVGSDGDD---- 79
+GGG GG + + + + NGG + G G GG GG D + D D
Sbjct: 144 KGGGGGGGET------ANKNKNKNKNNGGGNNKGKEGKKGGGGGGKLDKIDFDFQDFDIP 305
Query: 80 --GDGCGSGGGGGGLGGDIGSGSDGDDGGGGSGSGG 113
G G GGGGGG G G GG +G+GG
Sbjct: 306 SHGKSGGKGGGGGGKGNKNGHDHGHGIGGNMNGNGG 413
Score = 38.5 bits (88), Expect = 5e-04
Identities = 37/118 (31%), Positives = 50/118 (42%), Gaps = 32/118 (27%)
Frame = +3
Query: 27 EGGGCRGGNSNAEWWLGRRRRQRGVENGGD-------------DGGGGGG---------- 63
+GGG GG+S+++ ++++ +NGG GGGGGG
Sbjct: 36 KGGG--GGDSSSD-----KKKKTKDKNGGGFLVKFLGLGKKSKKGGGGGGGETANKNKNK 194
Query: 64 SGGGSGGDGVGSDGDDGDGCGSGGGGGGLGG--------DIGS-GSDGDDGGGGSGSG 112
+ GG+ G +G G GGGGG L DI S G G GGGG G G
Sbjct: 195 NKNNGGGNNKGKEGKKG-----GGGGGKLDKIDFDFQDFDIPSHGKSGGKGGGGGGKG 353
Score = 34.7 bits (78), Expect = 0.007
Identities = 20/54 (37%), Positives = 21/54 (38%)
Frame = +3
Query: 57 DGGGGGGSGGGSGGDGVGSDGDDGDGCGSGGGGGGLGGDIGSGSDGDDGGGGSG 110
DGG G GG G S D GGG L +G G GGGG G
Sbjct: 3 DGGNKSKEGKQKGGGGGDSSSDKKKKTKDKNGGGFLVKFLGLGKKSKKGGGGGG 164
>BP044109
Length = 497
Score = 46.6 bits (109), Expect = 2e-06
Identities = 42/119 (35%), Positives = 54/119 (45%), Gaps = 2/119 (1%)
Frame = -3
Query: 29 GGCRGGNSNAEWWLGRRRRQRGVENGGDDGGGGGGSGGGSGGDGVGSDGDDGDGCGSGGG 88
G RGG E + G GVE G ++ GG G G G + G G + +G +
Sbjct: 423 GVARGGEGRVEPFGGLGGD--GVEVGVEEDGGEGRVGAGPCEEEEGVSGGEVEGFVAKVD 250
Query: 89 GGGLGGDIGSGSDGDDGGG--GSGSGGSTVEVMLMMVVAVRVVVAATVEGGGCGGSDGR 145
GGGLG ++G DGGG G G +EV L A VV GGG GG +G+
Sbjct: 249 GGGLGDEVG------DGGGVVGVWVRGVDLEVAL---EACDGVVLFLFGGGGGGGGEGK 100
Score = 26.2 bits (56), Expect = 2.5
Identities = 24/70 (34%), Positives = 24/70 (34%), Gaps = 1/70 (1%)
Frame = -2
Query: 31 CRGGNSNAEWWLGRRRRQRGVENGGDDGGGGGGSGGGSGGDGVGSDGDDGDGCGSGGGG- 89
C W R R R E G G S GGS G G G G GGG
Sbjct: 463 CHPARRGQRWRRCRGRAWRRREGGAIWRARRGRSRGGS*GGWRGGKGWSRAM*GGGGGFR 284
Query: 90 GGLGGDIGSG 99
G GG G G
Sbjct: 283 GRSGGFCGEG 254
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.314 0.144 0.451
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,479,040
Number of Sequences: 28460
Number of extensions: 58221
Number of successful extensions: 6936
Number of sequences better than 10.0: 1032
Number of HSP's better than 10.0 without gapping: 1985
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 4308
length of query: 150
length of database: 4,897,600
effective HSP length: 82
effective length of query: 68
effective length of database: 2,563,880
effective search space: 174343840
effective search space used: 174343840
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 51 (24.3 bits)
Lotus: description of TM0090a.1


