

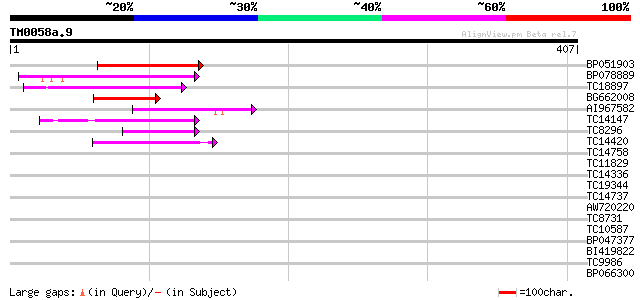
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0058a.9
(407 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BP051903 87 5e-18
BP078889 85 2e-17
TC18897 weakly similar to UP|Q9XEV3 (Q9XEV3) TIA-1 related prote... 79 2e-15
BG662008 57 6e-09
AI967582 54 5e-08
TC14147 similar to UP|ROC1_NICSY (Q08935) 29 kDa ribonucleoprote... 45 2e-05
TC8296 similar to GB|AAN28790.1|23505861|AY143851 At5g64200/MSJ1... 42 1e-04
TC14420 similar to UP|ROC3_NICSY (P19682) 28 kDa ribonucleoprote... 40 5e-04
TC14758 similar to UP|O24601 (O24601) Glycine-rich RNA binding p... 39 0.002
TC11829 weakly similar to UP|NAM8_YEAST (Q00539) NAM8 protein, p... 38 0.003
TC14336 similar to UP|Q9M549 (Q9M549) Poly(A)-binding protein, p... 37 0.005
TC19344 weakly similar to GB|AAA34838.1|172092|YSCPABPG polyaden... 37 0.008
TC14737 similar to UP|Q9LEB3 (Q9LEB3) RNA binding protein 47, pa... 36 0.010
AW720220 35 0.017
TC8731 homologue to UP|Q9LNS1 (Q9LNS1) F1L3.2, partial (34%) 35 0.017
TC10587 weakly similar to GB|AAC50895.1|1518802|HSU63289 CUG-BP/... 35 0.017
BP047377 35 0.023
BI419822 35 0.023
TC9986 similar to UP|Q9M427 (Q9M427) Oligouridylate binding prot... 35 0.030
BP066300 34 0.039
>BP051903
Length = 342
Score = 87.0 bits (214), Expect = 5e-18
Identities = 43/76 (56%), Positives = 54/76 (70%)
Frame = +2
Query: 64 IDGIEESFDYLHLRDICSKFGKVVNLFVQRQRKSGRFFRFGFFRYGLRDQALKAIQGLNG 123
+DGI + DY +R + ++FG+VVN+FVQRQRK GR FRFGF R+ + A KAI LNG
Sbjct: 68 VDGIPDRTDYYQIRGLFAQFGRVVNVFVQRQRKIGRLFRFGFVRFASLEVASKAIDFLNG 247
Query: 124 FRLGSAPLSASLAWAP 139
FRLG A LS S+A P
Sbjct: 248 FRLGDAALSVSMARFP 295
>BP078889
Length = 474
Score = 85.1 bits (209), Expect = 2e-17
Identities = 56/141 (39%), Positives = 79/141 (55%), Gaps = 11/141 (7%)
Frame = +1
Query: 7 ASRSFAPISAAARQIG------STTARG--VGDVAGEE---EGGELDGEADGGSRSCSAG 55
A+RS A A A ++ S A G + D+ GEE + GE +A G +R+
Sbjct: 46 AARSEAEQGATAEEVDEKDEVESEGALGGPIRDLGGEEVASDKGETRNDAGGLNRNNGGE 225
Query: 56 RRSCFSVFIDGIEESFDYLHLRDICSKFGKVVNLFVQRQRKSGRFFRFGFFRYGLRDQAL 115
RR + F+DGIEE+ +Y HLR + + FG++ NLFVQR+ K GR FRFGF R+ +QA
Sbjct: 226 RRRKPTPFVDGIEEAIEYHHLRGLFALFGRISNLFVQRKHKFGRRFRFGFVRFFSEEQAA 405
Query: 116 KAIQGLNGFRLGSAPLSASLA 136
L+G R+G LS + A
Sbjct: 406 AVAFVLDGLRVGGTALSVAPA 468
>TC18897 weakly similar to UP|Q9XEV3 (Q9XEV3) TIA-1 related protein, partial
(5%)
Length = 481
Score = 78.6 bits (192), Expect = 2e-15
Identities = 43/117 (36%), Positives = 64/117 (53%)
Frame = +1
Query: 11 FAPISAAARQIGSTTARGVGDVAGEEEGGELDGEADGGSRSCSAGRRSCFSVFIDGIEES 70
F P+S+ +GS V +G + +L + G + GR F++FIDGI E
Sbjct: 133 FLPLSSTRSILGSRRY-SVSGCSGVDSDEDLLEDEAGEEPAIGGGRAPSFTLFIDGISEP 309
Query: 71 FDYLHLRDICSKFGKVVNLFVQRQRKSGRFFRFGFFRYGLRDQALKAIQGLNGFRLG 127
+Y H+R + + G ++N+F+Q+QR+ R FRFGF RY RD A AI NG +G
Sbjct: 310 TNYYHIRGLFGQIGSLLNVFLQKQRRIRRNFRFGFVRYSTRDVASMAINLFNGVLMG 480
>BG662008
Length = 457
Score = 57.0 bits (136), Expect = 6e-09
Identities = 24/48 (50%), Positives = 35/48 (72%)
Frame = +3
Query: 61 SVFIDGIEESFDYLHLRDICSKFGKVVNLFVQRQRKSGRFFRFGFFRY 108
++F+DGI + DY +R + S G ++N+FVQRQR+ GR FRFGF R+
Sbjct: 102 TLFVDGISDFVDYSQIRGLFSHIGSLLNVFVQRQRRIGRAFRFGFVRF 245
>AI967582
Length = 356
Score = 53.9 bits (128), Expect = 5e-08
Identities = 35/104 (33%), Positives = 52/104 (49%), Gaps = 15/104 (14%)
Frame = +2
Query: 89 LFVQRQRKSGRFFRFGFFRYGLRDQALKAIQGLNGFRLGSAPLSASLAWAPWVCGSPR-- 146
+FVQR+ + GR F FGF + A A+ LNG ++G A LS +++ P G+P
Sbjct: 20 VFVQRKHEVGRRFHFGFVHFLSWSDAXTAVARLNGMKIGGAHLSVTVSTFPSRSGAPSGN 199
Query: 147 ---RGFGL----------CSQASSAQGQRLAARVLSWRDAVLGQ 177
R GL SQ+S +G + + SWRD V+G+
Sbjct: 200 SGFRALGLSKSD*RGSISVSQSSMLKGSVFPSTLPSWRDVVVGK 331
>TC14147 similar to UP|ROC1_NICSY (Q08935) 29 kDa ribonucleoprotein A,
chloroplast precursor (CP29A), partial (63%)
Length = 1602
Score = 45.1 bits (105), Expect = 2e-05
Identities = 37/115 (32%), Positives = 50/115 (43%)
Frame = +3
Query: 22 GSTTARGVGDVAGEEEGGELDGEADGGSRSCSAGRRSCFSVFIDGIEESFDYLHLRDICS 81
GS + RG D GG DG GGS S S R V + + D L +
Sbjct: 648 GSDSYRGGSD---GYRGGGSDGYRGGGSSSYSENR-----VHVGNLAWGVDNAALESLFR 803
Query: 82 KFGKVVNLFVQRQRKSGRFFRFGFFRYGLRDQALKAIQGLNGFRLGSAPLSASLA 136
+ G+VV+ V R+SGR FGF + D+ AI+ L+G L + S A
Sbjct: 804 EQGRVVDAKVIYDRESGRSRGFGFVTFSSPDEVNSAIRSLDGADLNGRAIKVSQA 968
>TC8296 similar to GB|AAN28790.1|23505861|AY143851 At5g64200/MSJ1_4
{Arabidopsis thaliana;}, partial (55%)
Length = 1276
Score = 42.4 bits (98), Expect = 1e-04
Identities = 19/55 (34%), Positives = 32/55 (57%)
Frame = +2
Query: 82 KFGKVVNLFVQRQRKSGRFFRFGFFRYGLRDQALKAIQGLNGFRLGSAPLSASLA 136
K+GKVV++F+ + R++G F F RY D+A KA+ L+G + ++ A
Sbjct: 197 KYGKVVDIFIPKDRRTGESRGFAFVRYKYADEASKAVDRLDGRMVDGREITVQFA 361
>TC14420 similar to UP|ROC3_NICSY (P19682) 28 kDa ribonucleoprotein,
chloroplast precursor (28RNP), partial (34%)
Length = 637
Score = 40.4 bits (93), Expect = 5e-04
Identities = 28/90 (31%), Positives = 41/90 (45%)
Frame = +1
Query: 60 FSVFIDGIEESFDYLHLRDICSKFGKVVNLFVQRQRKSGRFFRFGFFRYGLRDQALKAIQ 119
F V++ + FD L + S+ GKVV+ V R++GR FGF + AI
Sbjct: 40 FRVYVGNLPWDFDNSQLEQVFSEHGKVVSARVVYDRETGRSRGFGFVTMSDEAELNDAIA 219
Query: 120 GLNGFRLGSAPLSASLAWAPWVCGSPRRGF 149
L+G LG + ++A PRR F
Sbjct: 220 ALDGQSLGGRTIRVNVAE-----DRPRRSF 294
>TC14758 similar to UP|O24601 (O24601) Glycine-rich RNA binding protein 2,
partial (93%)
Length = 725
Score = 38.5 bits (88), Expect = 0.002
Identities = 21/79 (26%), Positives = 39/79 (48%)
Frame = +1
Query: 60 FSVFIDGIEESFDYLHLRDICSKFGKVVNLFVQRQRKSGRFFRFGFFRYGLRDQALKAIQ 119
+ F+ G+ + D L S +G++++ V R++GR FGF + + AI+
Sbjct: 82 YRCFVGGLAWTTDSHTLEQAFSSYGEIIDSKVVNDRETGRSRGFGFVTFTSEEAMRSAIE 261
Query: 120 GLNGFRLGSAPLSASLAWA 138
G+NG L ++ + A A
Sbjct: 262 GMNGNELDGRNITVNEAQA 318
>TC11829 weakly similar to UP|NAM8_YEAST (Q00539) NAM8 protein, partial (4%)
Length = 738
Score = 38.1 bits (87), Expect = 0.003
Identities = 30/132 (22%), Positives = 57/132 (42%), Gaps = 6/132 (4%)
Frame = +1
Query: 14 ISAAARQIGSTTARGVGDVAGEEEGGELDGEADGGSRSCSAGRRSC------FSVFIDGI 67
+ +A + ST A G G+ +G + GG+ G + +++I +
Sbjct: 61 VPESATKQTSTLAIGSGNSGSNPPWANNNGGSTGGAAQAGLGAGAVKKEIDDTNLYIGYL 240
Query: 68 EESFDYLHLRDICSKFGKVVNLFVQRQRKSGRFFRFGFFRYGLRDQALKAIQGLNGFRLG 127
+ + L + +FG++V V + R +G +GF +Y A AI +NG+RL
Sbjct: 241 PPTLEDDGLIQLFQQFGEIVMAKVIKDRMTGLSKGYGFVKYADITMANNAILAMNGYRLE 420
Query: 128 SAPLSASLAWAP 139
++ +A P
Sbjct: 421 GRTIAVRVAGKP 456
>TC14336 similar to UP|Q9M549 (Q9M549) Poly(A)-binding protein, partial
(85%)
Length = 1917
Score = 37.4 bits (85), Expect = 0.005
Identities = 22/77 (28%), Positives = 39/77 (50%)
Frame = +2
Query: 50 RSCSAGRRSCFSVFIDGIEESFDYLHLRDICSKFGKVVNLFVQRQRKSGRFFRFGFFRYG 109
R S + ++FI ++++ D+ L D S FG +++ V SG+ +GF ++
Sbjct: 62 RDPSIRKSGAGNIFIKNLDKAIDHKALHDTFSTFGNILSCKVATD-SSGQSKGYGFVQFE 238
Query: 110 LRDQALKAIQGLNGFRL 126
+ A AI+ LNG L
Sbjct: 239 TEEAAQNAIEKLNGMLL 289
Score = 37.0 bits (84), Expect = 0.006
Identities = 21/74 (28%), Positives = 36/74 (48%)
Frame = +2
Query: 50 RSCSAGRRSCFSVFIDGIEESFDYLHLRDICSKFGKVVNLFVQRQRKSGRFFRFGFFRYG 109
R S + +VF+ + ES L+++ +FG + + V R G+ FGF +
Sbjct: 335 RESSTDKAKFNNVFVKNLSESTTDDELKNVFGEFGTITSAVVMRDG-DGKSKCFGFVNFE 511
Query: 110 LRDQALKAIQGLNG 123
D A +A++ LNG
Sbjct: 512 NADDAARAVESLNG 553
>TC19344 weakly similar to GB|AAA34838.1|172092|YSCPABPG
polyadenylate-binding protein {Saccharomyces
cerevisiae;} , partial (18%)
Length = 518
Score = 36.6 bits (83), Expect = 0.008
Identities = 18/47 (38%), Positives = 29/47 (61%)
Frame = +2
Query: 81 SKFGKVVNLFVQRQRKSGRFFRFGFFRYGLRDQALKAIQGLNGFRLG 127
S FGK+V+L + + ++G FGF Y D A +A++ +NG +LG
Sbjct: 14 SSFGKIVSLTISKD-ENGMSKGFGFVNYENSDDAKRALEAMNGTQLG 151
>TC14737 similar to UP|Q9LEB3 (Q9LEB3) RNA binding protein 47, partial (32%)
Length = 803
Score = 36.2 bits (82), Expect = 0.010
Identities = 20/86 (23%), Positives = 42/86 (48%)
Frame = +3
Query: 61 SVFIDGIEESFDYLHLRDICSKFGKVVNLFVQRQRKSGRFFRFGFFRYGLRDQALKAIQG 120
++++ + D ++ + + G+VV+ V R +++G+ +GF + R A K +Q
Sbjct: 447 TIWLGDLHHWMDENYVHNCFAHTGEVVSAKVIRNKQTGQSEGYGFVEFYSRATAEKVLQN 626
Query: 121 LNGFRLGSAPLSASLAWAPWVCGSPR 146
NG + + + L WA + G R
Sbjct: 627 FNGTMMPNTDQAFRLNWASFSAGERR 704
>AW720220
Length = 535
Score = 35.4 bits (80), Expect = 0.017
Identities = 25/81 (30%), Positives = 38/81 (46%)
Frame = +3
Query: 58 SCFSVFIDGIEESFDYLHLRDICSKFGKVVNLFVQRQRKSGRFFRFGFFRYGLRDQALKA 117
+C SV++ I L L+++ + G V + R+ KS +GF Y R A A
Sbjct: 240 TCRSVYVGNIHTQVTELLLQEVFAGTGPVEGCKLFRKEKSS----YGFIHYFDRRSAALA 407
Query: 118 IQGLNGFRLGSAPLSASLAWA 138
I LNG L P+ + A+A
Sbjct: 408 ILTLNGRHLFGQPIKVNWAYA 470
>TC8731 homologue to UP|Q9LNS1 (Q9LNS1) F1L3.2, partial (34%)
Length = 721
Score = 35.4 bits (80), Expect = 0.017
Identities = 25/81 (30%), Positives = 38/81 (46%)
Frame = +2
Query: 58 SCFSVFIDGIEESFDYLHLRDICSKFGKVVNLFVQRQRKSGRFFRFGFFRYGLRDQALKA 117
+C SV++ I L L+++ + G V + R+ KS +GF Y R A A
Sbjct: 275 TCRSVYVGNIHTQVTELLLQEVFAGTGPVEGCKLFRKEKSS----YGFIHYFDRRSAALA 442
Query: 118 IQGLNGFRLGSAPLSASLAWA 138
I LNG L P+ + A+A
Sbjct: 443 ILTLNGRHLFGQPIKVNWAYA 505
>TC10587 weakly similar to GB|AAC50895.1|1518802|HSU63289 CUG-BP/hNab50
{Homo sapiens;} , partial (19%)
Length = 482
Score = 35.4 bits (80), Expect = 0.017
Identities = 22/71 (30%), Positives = 35/71 (48%)
Frame = +1
Query: 61 SVFIDGIEESFDYLHLRDICSKFGKVVNLFVQRQRKSGRFFRFGFFRYGLRDQALKAIQG 120
++FI I + F L + FG+V++ V + +G FGF Y + A AI
Sbjct: 16 NLFIYHIPQEFGDQDLANAFQPFGRVISAKVFVDKATGVSKCFGFVSYDSPEAAQSAISM 195
Query: 121 LNGFRLGSAPL 131
+NG++LG L
Sbjct: 196 MNGYQLGGKKL 228
>BP047377
Length = 564
Score = 35.0 bits (79), Expect = 0.023
Identities = 21/75 (28%), Positives = 34/75 (45%)
Frame = +3
Query: 62 VFIDGIEESFDYLHLRDICSKFGKVVNLFVQRQRKSGRFFRFGFFRYGLRDQALKAIQGL 121
VF+ I L +IC + G VV+ + R++G+ +GF Y + AL A + L
Sbjct: 135 VFVGNIPYDATEEQLIEICQEVGPVVSFRLVIDRETGKPKGYGFCEYKDEETALSARRNL 314
Query: 122 NGFRLGSAPLSASLA 136
G+ + L A
Sbjct: 315 QGYEINGRQLRVDFA 359
>BI419822
Length = 380
Score = 35.0 bits (79), Expect = 0.023
Identities = 21/77 (27%), Positives = 35/77 (45%)
Frame = +2
Query: 60 FSVFIDGIEESFDYLHLRDICSKFGKVVNLFVQRQRKSGRFFRFGFFRYGLRDQALKAIQ 119
F F+ G+ + D L S +G++V V R++GR FGF + AI+
Sbjct: 77 FRCFVGGLAWATDNEALEKAFSPYGEIVESKVINDRETGRSRGFGFVTFASEQAMKDAIE 256
Query: 120 GLNGFRLGSAPLSASLA 136
+NG L ++ + A
Sbjct: 257 AMNGQNLDGRNITVNEA 307
>TC9986 similar to UP|Q9M427 (Q9M427) Oligouridylate binding protein,
partial (46%)
Length = 1150
Score = 34.7 bits (78), Expect = 0.030
Identities = 45/185 (24%), Positives = 64/185 (34%), Gaps = 20/185 (10%)
Frame = +3
Query: 19 RQIGSTTA-RGVGDVAGEEEG-----------GELDGEADGGSRSCSAGRRSCFSVFIDG 66
RQI A +G G + EE+ G DG D + S +V++
Sbjct: 192 RQIRCNWATKGAGGSSSEEKNSDNQNAVVLTNGSSDGGQDNNNEEAPENNPSYTTVYVGN 371
Query: 67 IEESFDYLHLRDICSKFGKVVNLFVQRQRKSGRFFRFGFFRYGLRDQALKAIQGLNGFRL 126
+ L G V V+ QR G FGF RY ++A AIQ NG +
Sbjct: 372 LPHDVTQAELHCQFHALGAGVIEEVRIQRDKG----FGFVRYNTHEEAALAIQVANGRIV 539
Query: 127 GSAPLSASLAWAPWVCGS--------PRRGFGLCSQASSAQGQRLAARVLSWRDAVLGQR 178
+ S P G+ + + + A QG A + R L Q
Sbjct: 540 RGKTMKCSWGSRPTPPGTASNPLPPPAAQPYQILPTAGMNQGYSPAELLAYQRQLALSQA 719
Query: 179 KVSGV 183
VSG+
Sbjct: 720 AVSGL 734
>BP066300
Length = 420
Score = 34.3 bits (77), Expect = 0.039
Identities = 15/48 (31%), Positives = 29/48 (60%)
Frame = -3
Query: 76 LRDICSKFGKVVNLFVQRQRKSGRFFRFGFFRYGLRDQALKAIQGLNG 123
LR+ FG++V++ + + + R F F RY +++ KAI+G++G
Sbjct: 385 LRNCFKNFGQLVDVXLVMDKLANRPRGFAFLRYATEEESQKAIEGMHG 242
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.325 0.142 0.455
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,238,334
Number of Sequences: 28460
Number of extensions: 138070
Number of successful extensions: 1386
Number of sequences better than 10.0: 74
Number of HSP's better than 10.0 without gapping: 1364
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1380
length of query: 407
length of database: 4,897,600
effective HSP length: 92
effective length of query: 315
effective length of database: 2,279,280
effective search space: 717973200
effective search space used: 717973200
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 56 (26.2 bits)
Lotus: description of TM0058a.9


