

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0058a.3
(579 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
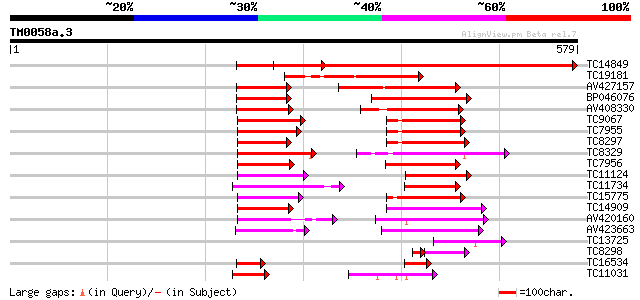
Score E
Sequences producing significant alignments: (bits) Value
TC14849 similar to UP|Q40090 (Q40090) SPF1 protein, partial (37%) 645 0.0
TC19181 similar to UP|Q40090 (Q40090) SPF1 protein, partial (19%) 207 3e-54
AV427157 164 4e-41
BP046076 123 9e-29
AV408330 104 3e-23
TC9067 similar to UP|WR15_ARATH (O22176) Probable WRKY transcrip... 92 2e-19
TC7955 similar to UP|WR11_ARATH (Q9SV15) Probable WRKY transcrip... 92 2e-19
TC8297 similar to UP|Q40828 (Q40828) WRKY3, partial (27%) 91 7e-19
TC8329 similar to UP|BAD06717 (BAD06717) WRKY transcription fact... 90 1e-18
TC7956 similar to GB|AAM61419.1|21537078|AY084855 putaive DNA-bi... 88 3e-18
TC11124 similar to UP|Q8H9D7 (Q8H9D7) WRKY-type DNA binding prot... 87 6e-18
TC11734 similar to UP|WR49_ARATH (Q9FHR7) Probable WRKY transcri... 87 6e-18
TC15775 similar to UP|WR17_ARATH (Q9SJA8) Probable WRKY transcri... 87 7e-18
TC14909 similar to UP|WR21_ARATH (O04336) Probable WRKY transcri... 84 6e-17
AV420160 77 6e-15
AV423663 75 2e-14
TC13725 similar to UP|Q39658 (Q39658) SPF1-like DNA-binding prot... 72 2e-13
TC8298 homologue to UP|Q9SXP4 (Q9SXP4) DNA-binding protein NtWRK... 53 3e-09
TC16534 similar to UP|WR41_ARATH (Q8H0Y8) Probable WRKY transcri... 57 1e-08
TC11031 homologue to UP|WR65_ARATH (Q9LP56) Probable WRKY transc... 53 2e-07
>TC14849 similar to UP|Q40090 (Q40090) SPF1 protein, partial (37%)
Length = 1389
Score = 645 bits (1664), Expect = 0.0
Identities = 310/310 (100%), Positives = 310/310 (100%)
Frame = +2
Query: 270 RSLDGQITEIVYKGTHNHPKPQATRRNSSSSSSLPIPHSNHISNDIQDQSYATHGSGQMD 329
RSLDGQITEIVYKGTHNHPKPQATRRNSSSSSSLPIPHSNHISNDIQDQSYATHGSGQMD
Sbjct: 2 RSLDGQITEIVYKGTHNHPKPQATRRNSSSSSSLPIPHSNHISNDIQDQSYATHGSGQMD 181
Query: 330 SVATPENSSISIGDDDFEQSSQKCRSGGDELDEDEPDSKRWKVEGENEGISAPGSRTVRE 389
SVATPENSSISIGDDDFEQSSQKCRSGGDELDEDEPDSKRWKVEGENEGISAPGSRTVRE
Sbjct: 182 SVATPENSSISIGDDDFEQSSQKCRSGGDELDEDEPDSKRWKVEGENEGISAPGSRTVRE 361
Query: 390 PRVVVQTTSDIDILDDGYRWRKYGQKVVKGNPNPRSYYKCTHPGCPVRKHVERASHDLRA 449
PRVVVQTTSDIDILDDGYRWRKYGQKVVKGNPNPRSYYKCTHPGCPVRKHVERASHDLRA
Sbjct: 362 PRVVVQTTSDIDILDDGYRWRKYGQKVVKGNPNPRSYYKCTHPGCPVRKHVERASHDLRA 541
Query: 450 VITTYEGKHNHDVPAARGSGNHSVTRPLPNNTANAIRPSSVTHQHHHNTNNNSLQSLRPQ 509
VITTYEGKHNHDVPAARGSGNHSVTRPLPNNTANAIRPSSVTHQHHHNTNNNSLQSLRPQ
Sbjct: 542 VITTYEGKHNHDVPAARGSGNHSVTRPLPNNTANAIRPSSVTHQHHHNTNNNSLQSLRPQ 721
Query: 510 QAPDQVQSSPFTLEMLQQNQGGFGFSSGFGNSMGSYNMNNQQQQQLPDNVFSSRAKEEPR 569
QAPDQVQSSPFTLEMLQQNQGGFGFSSGFGNSMGSYNMNNQQQQQLPDNVFSSRAKEEPR
Sbjct: 722 QAPDQVQSSPFTLEMLQQNQGGFGFSSGFGNSMGSYNMNNQQQQQLPDNVFSSRAKEEPR 901
Query: 570 DDTFFDSLLC 579
DDTFFDSLLC
Sbjct: 902 DDTFFDSLLC 931
Score = 92.4 bits (228), Expect = 2e-19
Identities = 49/93 (52%), Positives = 61/93 (64%), Gaps = 1/93 (1%)
Frame = +2
Query: 232 DDGYNWRKYGQKQVKGSENPRSYYKCTYPNCPTKKKVER-SLDGQITEIVYKGTHNHPKP 290
DDGY WRKYGQK VKG+ NPRSYYKCT+P CP +K VER S D + Y+G HNH P
Sbjct: 404 DDGYRWRKYGQKVVKGNPNPRSYYKCTHPGCPVRKHVERASHDLRAVITTYEGKHNHDVP 583
Query: 291 QATRRNSSSSSSLPIPHSNHISNDIQDQSYATH 323
A R + + S + P+P N+ +N I+ S TH
Sbjct: 584 -AARGSGNHSVTRPLP--NNTANAIRPSS-VTH 670
>TC19181 similar to UP|Q40090 (Q40090) SPF1 protein, partial (19%)
Length = 402
Score = 207 bits (528), Expect = 3e-54
Identities = 104/142 (73%), Positives = 119/142 (83%)
Frame = +3
Query: 281 YKGTHNHPKPQATRRNSSSSSSLPIPHSNHISNDIQDQSYATHGSGQMDSVATPENSSIS 340
YKGTHNHPKPQA + +SS ++ SNH+S +IQ ATH SGQMDSVATPENSS+S
Sbjct: 3 YKGTHNHPKPQAPHKRNSSFTT-----SNHVSTEIQG---ATHVSGQMDSVATPENSSLS 158
Query: 341 IGDDDFEQSSQKCRSGGDELDEDEPDSKRWKVEGENEGISAPGSRTVREPRVVVQTTSDI 400
+G DDFEQSS +SGG+ELDEDEP K+W++EGENEG+SA GSRTV+E RVVVQTTSDI
Sbjct: 159 MGGDDFEQSSMS-KSGGEELDEDEPHGKKWRMEGENEGMSAAGSRTVKESRVVVQTTSDI 335
Query: 401 DILDDGYRWRKYGQKVVKGNPN 422
DILDDGYRWRKYGQKVVKGNPN
Sbjct: 336 DILDDGYRWRKYGQKVVKGNPN 401
Score = 38.5 bits (88), Expect = 0.003
Identities = 15/19 (78%), Positives = 16/19 (83%)
Frame = +3
Query: 232 DDGYNWRKYGQKQVKGSEN 250
DDGY WRKYGQK VKG+ N
Sbjct: 345 DDGYRWRKYGQKVVKGNPN 401
>AV427157
Length = 379
Score = 164 bits (415), Expect = 4e-41
Identities = 80/125 (64%), Positives = 99/125 (79%)
Frame = +3
Query: 336 NSSISIGDDDFEQSSQKCRSGGDELDEDEPDSKRWKVEGENEGISAPGSRTVREPRVVVQ 395
+S+ S +++ +Q + S G + + DE +SKR K+E +S +R +REPRVVVQ
Sbjct: 6 SSTFSNEEEEDDQGTHGSVSLGYDGEGDESESKRRKLESYATELSG-ATRAIREPRVVVQ 182
Query: 396 TTSDIDILDDGYRWRKYGQKVVKGNPNPRSYYKCTHPGCPVRKHVERASHDLRAVITTYE 455
TTS++DILDDGYRWRKYGQKVVKGNPNPRSYYKCT+ GC VRKHVERASHDL++VITTYE
Sbjct: 183 TTSEVDILDDGYRWRKYGQKVVKGNPNPRSYYKCTNAGCMVRKHVERASHDLKSVITTYE 362
Query: 456 GKHNH 460
GKHNH
Sbjct: 363 GKHNH 377
Score = 74.7 bits (182), Expect = 4e-14
Identities = 35/57 (61%), Positives = 39/57 (68%), Gaps = 1/57 (1%)
Frame = +3
Query: 232 DDGYNWRKYGQKQVKGSENPRSYYKCTYPNCPTKKKVER-SLDGQITEIVYKGTHNH 287
DDGY WRKYGQK VKG+ NPRSYYKCT C +K VER S D + Y+G HNH
Sbjct: 207 DDGYRWRKYGQKVVKGNPNPRSYYKCTNAGCMVRKHVERASHDLKSVITTYEGKHNH 377
>BP046076
Length = 593
Score = 123 bits (308), Expect = 9e-29
Identities = 57/102 (55%), Positives = 67/102 (64%)
Frame = -2
Query: 370 WKVEGENEGISAPGSRTVREPRVVVQTTSDIDILDDGYRWRKYGQKVVKGNPNPRSYYKC 429
W G +E R +REPR QT SD+D+LDDGY+WRKYGQKVVK + +PRSYY+C
Sbjct: 559 WWRSGGSEKSKVKVRRKLREPRFCFQTRSDVDVLDDGYKWRKYGQKVVKNSLHPRSYYRC 380
Query: 430 THPGCPVRKHVERASHDLRAVITTYEGKHNHDVPAARGSGNH 471
TH C V+K VER S D R VITTYEG+HNH S H
Sbjct: 379 THSNCRVKKRVERLSEDCRMVITTYEGRHNHSPCDESNSSEH 254
Score = 77.4 bits (189), Expect = 6e-15
Identities = 35/57 (61%), Positives = 42/57 (73%), Gaps = 1/57 (1%)
Frame = -2
Query: 232 DDGYNWRKYGQKQVKGSENPRSYYKCTYPNCPTKKKVER-SLDGQITEIVYKGTHNH 287
DDGY WRKYGQK VK S +PRSYY+CT+ NC KK+VER S D ++ Y+G HNH
Sbjct: 457 DDGYKWRKYGQKVVKNSLHPRSYYRCTHSNCRVKKRVERLSEDCRMVITTYEGRHNH 287
>AV408330
Length = 316
Score = 104 bits (260), Expect = 3e-23
Identities = 51/105 (48%), Positives = 64/105 (60%)
Frame = +3
Query: 359 ELDEDEPDSKRWKVEGENEGISAPGSRTVREPRVVVQTTSDIDILDDGYRWRKYGQKVVK 418
E DE K+ K + N+ + REPR T S++D L+DGYRWRKYGQK VK
Sbjct: 15 EDDEHNKTKKQLKAKKTNQ-------KRQREPRFAFMTKSEVDHLEDGYRWRKYGQKAVK 173
Query: 419 GNPNPRSYYKCTHPGCPVRKHVERASHDLRAVITTYEGKHNHDVP 463
+P PRSYY+CT C V+K VER+ D V+TTYEG+H H P
Sbjct: 174 NSPFPRSYYRCTSASCNVKKRVERSFTDPSIVVTTYEGQHTHSSP 308
Score = 74.7 bits (182), Expect = 4e-14
Identities = 34/60 (56%), Positives = 39/60 (64%), Gaps = 1/60 (1%)
Frame = +3
Query: 232 DDGYNWRKYGQKQVKGSENPRSYYKCTYPNCPTKKKVERSL-DGQITEIVYKGTHNHPKP 290
+DGY WRKYGQK VK S PRSYY+CT +C KK+VERS D I Y+G H H P
Sbjct: 129 EDGYRWRKYGQKAVKNSPFPRSYYRCTSASCNVKKRVERSFTDPSIVVTTYEGQHTHSSP 308
>TC9067 similar to UP|WR15_ARATH (O22176) Probable WRKY transcription
factor 15 (WRKY DNA-binding protein 15), partial (31%)
Length = 660
Score = 92.4 bits (228), Expect = 2e-19
Identities = 44/82 (53%), Positives = 55/82 (66%), Gaps = 1/82 (1%)
Frame = +3
Query: 385 RTVREPRVVVQTTSDIDILDDGYRWRKYGQKVVKGNPNPRSYYKCTH-PGCPVRKHVERA 443
R VR P + ++ DI D Y WRKYGQK +KG+P+PR YYKC+ GCP RKHVERA
Sbjct: 75 RVVRVPAISLKMA---DIPPDDYSWRKYGQKPIKGSPHPRGYYKCSSVRGCPARKHVERA 245
Query: 444 SHDLRAVITTYEGKHNHDVPAA 465
D ++ TYEG+HNH + AA
Sbjct: 246 LDDAAMLVVTYEGEHNHALSAA 311
Score = 71.6 bits (174), Expect = 3e-13
Identities = 35/72 (48%), Positives = 44/72 (60%), Gaps = 2/72 (2%)
Frame = +3
Query: 233 DGYNWRKYGQKQVKGSENPRSYYKC-TYPNCPTKKKVERSL-DGQITEIVYKGTHNHPKP 290
D Y+WRKYGQK +KGS +PR YYKC + CP +K VER+L D + + Y+G HNH
Sbjct: 126 DDYSWRKYGQKPIKGSPHPRGYYKCSSVRGCPARKHVERALDDAAMLVVTYEGEHNHALS 305
Query: 291 QATRRNSSSSSS 302
A N SS
Sbjct: 306 AADATNLILESS 341
>TC7955 similar to UP|WR11_ARATH (Q9SV15) Probable WRKY transcription
factor 11 (WRKY DNA-binding protein 11), partial (27%)
Length = 579
Score = 92.0 bits (227), Expect = 2e-19
Identities = 46/82 (56%), Positives = 55/82 (66%), Gaps = 1/82 (1%)
Frame = +3
Query: 385 RTVREPRVVVQTTSDIDILDDGYRWRKYGQKVVKGNPNPRSYYKC-THPGCPVRKHVERA 443
+TVR P + +T DI D Y WRKYGQK +KG+P PR YYKC T GCP RKHVERA
Sbjct: 42 KTVRVPAISSKTA---DIPPDEYSWRKYGQKPIKGSPYPRGYYKCSTVRGCPARKHVERA 212
Query: 444 SHDLRAVITTYEGKHNHDVPAA 465
+ D +I TYEG+H H + AA
Sbjct: 213 TDDPTMLIVTYEGEHRHALQAA 278
Score = 67.8 bits (164), Expect = 5e-12
Identities = 34/68 (50%), Positives = 41/68 (60%), Gaps = 2/68 (2%)
Frame = +3
Query: 233 DGYNWRKYGQKQVKGSENPRSYYKC-TYPNCPTKKKVERSLDGQITEIV-YKGTHNHPKP 290
D Y+WRKYGQK +KGS PR YYKC T CP +K VER+ D IV Y+G H H
Sbjct: 93 DEYSWRKYGQKPIKGSPYPRGYYKCSTVRGCPARKHVERATDDPTMLIVTYEGEHRHALQ 272
Query: 291 QATRRNSS 298
A ++S
Sbjct: 273 AAMHEDAS 296
>TC8297 similar to UP|Q40828 (Q40828) WRKY3, partial (27%)
Length = 617
Score = 90.5 bits (223), Expect = 7e-19
Identities = 43/86 (50%), Positives = 54/86 (62%), Gaps = 1/86 (1%)
Frame = +1
Query: 385 RTVREPRVVVQTTSDIDILDDGYRWRKYGQKVVKGNPNPRSYYKC-THPGCPVRKHVERA 443
RT+R P + +T DI D ++WRKYGQK +KG+P PR YYKC + GCP RKHVERA
Sbjct: 121 RTIRVPAISSKTA---DIPTDDFQWRKYGQKPIKGSPYPRGYYKCSSEKGCPARKHVERA 291
Query: 444 SHDLRAVITTYEGKHNHDVPAARGSG 469
D +I TYEG+H H + G
Sbjct: 292 QDDPNMLIVTYEGEHRHLITTTAAVG 369
Score = 62.0 bits (149), Expect = 3e-10
Identities = 28/57 (49%), Positives = 36/57 (63%), Gaps = 2/57 (3%)
Frame = +1
Query: 233 DGYNWRKYGQKQVKGSENPRSYYKC-TYPNCPTKKKVERSLDG-QITEIVYKGTHNH 287
D + WRKYGQK +KGS PR YYKC + CP +K VER+ D + + Y+G H H
Sbjct: 172 DDFQWRKYGQKPIKGSPYPRGYYKCSSEKGCPARKHVERAQDDPNMLIVTYEGEHRH 342
>TC8329 similar to UP|BAD06717 (BAD06717) WRKY transcription factor 1,
partial (41%)
Length = 1717
Score = 89.7 bits (221), Expect = 1e-18
Identities = 57/163 (34%), Positives = 89/163 (53%), Gaps = 7/163 (4%)
Frame = +1
Query: 355 SGGDELDEDEPDSKRWKVEGENEGISAPGSRTVREPRVVVQTTSDIDILDDGYRWRKYGQ 414
+G E + DS + + E I A SR V + T ++ DGY+WRKYGQ
Sbjct: 457 NGNSESSSTDEDSFK---KPREETIKAKISRAY----VRTEATDTGFVVKDGYQWRKYGQ 615
Query: 415 KVVKGNPNPRSYYKCTH-PGCPVRKHVERASHDLRAVITTYEGKHNHDVP----AARGSG 469
KV + NP PR+Y+KC+ P CPV+K V+R+ D ++ TYEG+HNH P A GSG
Sbjct: 616 KVTRDNPCPRAYFKCSFAPSCPVKKKVQRSVDDQSVLVATYEGEHNHPQPSQMEATSGSG 795
Query: 470 -NHSVTRPLPNN-TANAIRPSSVTHQHHHNTNNNSLQSLRPQQ 510
N S+ +P++ + ++ P+ VT + +N ++ P++
Sbjct: 796 RNVSLVGSMPSSKSLSSPAPAVVTLDLTKSRCSNDSKNAEPRK 924
Score = 80.9 bits (198), Expect = 5e-16
Identities = 38/87 (43%), Positives = 55/87 (62%), Gaps = 6/87 (6%)
Frame = +1
Query: 233 DGYNWRKYGQKQVKGSENPRSYYKCTY-PNCPTKKKVERSLDGQ-ITEIVYKGTHNHPKP 290
DGY WRKYGQK + + PR+Y+KC++ P+CP KKKV+RS+D Q + Y+G HNHP+P
Sbjct: 586 DGYQWRKYGQKVTRDNPCPRAYFKCSFAPSCPVKKKVQRSVDDQSVLVATYEGEHNHPQP 765
Query: 291 QATRRNSSSSSSLPI----PHSNHISN 313
S S ++ + P S +S+
Sbjct: 766 SQMEATSGSGRNVSLVGSMPSSKSLSS 846
>TC7956 similar to GB|AAM61419.1|21537078|AY084855 putaive DNA-binding
protein {Arabidopsis thaliana;} , partial (45%)
Length = 1257
Score = 88.2 bits (217), Expect = 3e-18
Identities = 43/78 (55%), Positives = 50/78 (63%), Gaps = 1/78 (1%)
Frame = +3
Query: 384 SRTVREPRVVVQTTSDIDILDDGYRWRKYGQKVVKGNPNPRSYYKC-THPGCPVRKHVER 442
SR + RV ++ DI D Y WRKYGQK +KG+P PR YYKC T GCP RKHVER
Sbjct: 591 SRVKQTIRVPAVSSKIADIPPDEYSWRKYGQKPIKGSPYPRGYYKCSTVRGCPARKHVER 770
Query: 443 ASHDLRAVITTYEGKHNH 460
A D +I TYEG+H H
Sbjct: 771 AQDDPNMLIVTYEGEHRH 824
Score = 66.6 bits (161), Expect = 1e-11
Identities = 31/61 (50%), Positives = 39/61 (63%), Gaps = 2/61 (3%)
Frame = +3
Query: 233 DGYNWRKYGQKQVKGSENPRSYYKC-TYPNCPTKKKVERSLDG-QITEIVYKGTHNHPKP 290
D Y+WRKYGQK +KGS PR YYKC T CP +K VER+ D + + Y+G H H +
Sbjct: 654 DEYSWRKYGQKPIKGSPYPRGYYKCSTVRGCPARKHVERAQDDPNMLIVTYEGEHRHQQQ 833
Query: 291 Q 291
Q
Sbjct: 834 Q 836
Score = 29.3 bits (64), Expect = 1.8
Identities = 22/53 (41%), Positives = 24/53 (44%)
Frame = +1
Query: 55 TGSGVPKFKSTPPPSLPLSPPTIFIPPGLSPAELLDSPVLLNSSNILPSPTTG 107
T S P TPPPS P SPPT P S L P S++ P P TG
Sbjct: 388 TPSPSPPPTPTPPPSNPPSPPTAASPTARSAPS*LQLP-----SHLSP-PLTG 528
>TC11124 similar to UP|Q8H9D7 (Q8H9D7) WRKY-type DNA binding protein,
partial (45%)
Length = 517
Score = 87.4 bits (215), Expect = 6e-18
Identities = 38/67 (56%), Positives = 44/67 (64%)
Frame = +1
Query: 405 DGYRWRKYGQKVVKGNPNPRSYYKCTHPGCPVRKHVERASHDLRAVITTYEGKHNHDVPA 464
DGYRWRKYGQK VK N PRSYY+CTH GC V+K V R + D V+TTYEG H H +
Sbjct: 1 DGYRWRKYGQKAVKNNKFPRSYYRCTHHGCHVKKPVPRLTQDEGVVVTTYEGVHTHPIEK 180
Query: 465 ARGSGNH 471
+ H
Sbjct: 181 TTDNFEH 201
Score = 70.9 bits (172), Expect = 5e-13
Identities = 33/74 (44%), Positives = 43/74 (57%), Gaps = 1/74 (1%)
Frame = +1
Query: 233 DGYNWRKYGQKQVKGSENPRSYYKCTYPNCPTKKKVER-SLDGQITEIVYKGTHNHPKPQ 291
DGY WRKYGQK VK ++ PRSYY+CT+ C KK V R + D + Y+G H HP +
Sbjct: 1 DGYRWRKYGQKAVKNNKFPRSYYRCTHHGCHVKKPVPRLTQDEGVVVTTYEGVHTHPIEK 180
Query: 292 ATRRNSSSSSSLPI 305
T + +PI
Sbjct: 181 TTDNFEHILNQMPI 222
>TC11734 similar to UP|WR49_ARATH (Q9FHR7) Probable WRKY transcription
factor 49 (WRKY DNA-binding protein 49), partial (28%)
Length = 1009
Score = 87.4 bits (215), Expect = 6e-18
Identities = 35/57 (61%), Positives = 44/57 (76%)
Frame = +3
Query: 404 DDGYRWRKYGQKVVKGNPNPRSYYKCTHPGCPVRKHVERASHDLRAVITTYEGKHNH 460
DDGY+WRKYGQK +K +PNPRSYY+CT+P C +K VER++ D +I TYEG H H
Sbjct: 333 DDGYKWRKYGQKSIKNSPNPRSYYRCTNPRCSAKKQVERSNEDPETLIITYEGLHLH 503
Score = 84.7 bits (208), Expect = 4e-17
Identities = 48/118 (40%), Positives = 62/118 (51%), Gaps = 3/118 (2%)
Frame = +3
Query: 228 NRRSDDGYNWRKYGQKQVKGSENPRSYYKCTYPNCPTKKKVERSLDGQITEIV-YKGTHN 286
N DDGY WRKYGQK +K S NPRSYY+CT P C KK+VERS + T I+ Y+G H
Sbjct: 321 NGMGDDGYKWRKYGQKSIKNSPNPRSYYRCTNPRCSAKKQVERSNEDPETLIITYEGLHL 500
Query: 287 HPKPQA--TRRNSSSSSSLPIPHSNHISNDIQDQSYATHGSGQMDSVATPENSSISIG 342
H + S S P+ S S+ +Q Q+ A D + +S S+G
Sbjct: 501 HFAYPCFLVGQPQQSQSHPPVKKSKFTSSQVQAQAQA------QDFIVNEAQASASVG 656
>TC15775 similar to UP|WR17_ARATH (Q9SJA8) Probable WRKY transcription
factor 17 (WRKY DNA-binding protein 17), partial (53%)
Length = 1284
Score = 87.0 bits (214), Expect = 7e-18
Identities = 43/82 (52%), Positives = 53/82 (64%), Gaps = 1/82 (1%)
Frame = +3
Query: 385 RTVREPRVVVQTTSDIDILDDGYRWRKYGQKVVKGNPNPRSYYKC-THPGCPVRKHVERA 443
+T+R P V ++ DI D Y WRKYGQK +KG P PR YYKC T GCP RKHVERA
Sbjct: 690 KTIRVPAV---SSKVADIPADEYSWRKYGQKPIKGTPYPRGYYKCSTVRGCPARKHVERA 860
Query: 444 SHDLRAVITTYEGKHNHDVPAA 465
+ D +I TYE +H+H + A
Sbjct: 861 TDDPAMLIVTYEDEHDHGIQTA 926
Score = 63.9 bits (154), Expect = 7e-11
Identities = 33/70 (47%), Positives = 41/70 (58%), Gaps = 2/70 (2%)
Frame = +3
Query: 233 DGYNWRKYGQKQVKGSENPRSYYKC-TYPNCPTKKKVERSLDGQITEIV-YKGTHNHPKP 290
D Y+WRKYGQK +KG+ PR YYKC T CP +K VER+ D IV Y+ H+H
Sbjct: 741 DEYSWRKYGQKPIKGTPYPRGYYKCSTVRGCPARKHVERATDDPAMLIVTYEDEHDHGIQ 920
Query: 291 QATRRNSSSS 300
A N S +
Sbjct: 921 TAMHDNISGA 950
>TC14909 similar to UP|WR21_ARATH (O04336) Probable WRKY transcription
factor 21 (WRKY DNA-binding protein 21), partial (22%)
Length = 590
Score = 84.0 bits (206), Expect = 6e-17
Identities = 44/106 (41%), Positives = 60/106 (56%), Gaps = 3/106 (2%)
Frame = +3
Query: 385 RTVREPRVVVQTTSDIDILDDGYRWRKYGQKVVKGNPNPRSYYKCTH-PGCPVRKHVERA 443
R R +V + +I D Y WRKYGQK +KG+P+PR YYKC+ GCP RKHVER
Sbjct: 3 RVKRSIKVPAISNKLANIPPDDYSWRKYGQKPIKGSPHPRGYYKCSSMRGCPARKHVERC 182
Query: 444 SHDLRAVITTYEGKHNHDVPAARGSGNHSVTR--PLPNNTANAIRP 487
+ ++ TYEG+HNH + + +V R P P + N +P
Sbjct: 183 LEEPTMLLVTYEGEHNHPKIPTQPANA*TVWRA*PRPGPSFNLSKP 320
Score = 71.6 bits (174), Expect = 3e-13
Identities = 33/59 (55%), Positives = 41/59 (68%), Gaps = 2/59 (3%)
Frame = +3
Query: 233 DGYNWRKYGQKQVKGSENPRSYYKC-TYPNCPTKKKVERSLDGQITEIV-YKGTHNHPK 289
D Y+WRKYGQK +KGS +PR YYKC + CP +K VER L+ +V Y+G HNHPK
Sbjct: 63 DDYSWRKYGQKPIKGSPHPRGYYKCSSMRGCPARKHVERCLEEPTMLLVTYEGEHNHPK 239
>AV420160
Length = 417
Score = 77.4 bits (189), Expect = 6e-15
Identities = 44/128 (34%), Positives = 64/128 (49%), Gaps = 12/128 (9%)
Frame = +1
Query: 374 GENEGISAPGSRTVRE--PRVVVQTTSDIDIL---------DDGYRWRKYGQKVVKGNPN 422
GE+ AP + RE RVV D+D D + WRKYGQK +KG+P
Sbjct: 31 GEDIKTEAPSPKKRREMKKRVVTLPIGDVDGSKSKGETYPPSDSWAWRKYGQKPIKGSPY 210
Query: 423 PRSYYKC-THPGCPVRKHVERASHDLRAVITTYEGKHNHDVPAARGSGNHSVTRPLPNNT 481
PR YY+C + GCP RK VER+ D ++ TY +HNH +P + + + ++
Sbjct: 211 PRGYYRCSSSKGCPARKQVERSRVDPTKLMVTYSYEHNHSLPVPKSHSSAASATATTSSD 390
Query: 482 ANAIRPSS 489
A P++
Sbjct: 391 GAADTPAT 414
Score = 65.9 bits (159), Expect = 2e-11
Identities = 39/104 (37%), Positives = 52/104 (49%), Gaps = 2/104 (1%)
Frame = +1
Query: 233 DGYNWRKYGQKQVKGSENPRSYYKC-TYPNCPTKKKVERS-LDGQITEIVYKGTHNHPKP 290
D + WRKYGQK +KGS PR YY+C + CP +K+VERS +D + Y HNH
Sbjct: 157 DSWAWRKYGQKPIKGSPYPRGYYRCSSSKGCPARKQVERSRVDPTKLMVTYSYEHNH--- 327
Query: 291 QATRRNSSSSSSLPIPHSNHISNDIQDQSYATHGSGQMDSVATP 334
SLP+P S+ + + T G D+ ATP
Sbjct: 328 -----------SLPVPKSH---SSAASATATTSSDGAADTPATP 417
>AV423663
Length = 447
Score = 75.5 bits (184), Expect = 2e-14
Identities = 42/107 (39%), Positives = 56/107 (52%), Gaps = 2/107 (1%)
Frame = +1
Query: 380 SAPGSRTVREPRVVVQTTSDIDILDDGYRWRKYGQKVVKGNPNPRSYYKC-THPGCPVRK 438
+ P S+ + V S ++ D + WRKYGQK +KG+P PR YY+C + GC RK
Sbjct: 7 TTPRSKRRKNQFKKVCQVSAENLSSDIWAWRKYGQKPIKGSPYPRGYYRCSSSKGCLARK 186
Query: 439 HVERASHDLRAVITTYEGKHNHDVPAARGS-GNHSVTRPLPNNTANA 484
VER + I TY G+HNH P R S + +PL TA A
Sbjct: 187 QVERNKSEPAMFIVTYTGEHNHPAPTHRNSLAGSTRQKPLTPETATA 327
Score = 66.2 bits (160), Expect = 1e-11
Identities = 37/78 (47%), Positives = 46/78 (58%), Gaps = 2/78 (2%)
Frame = +1
Query: 231 SDDGYNWRKYGQKQVKGSENPRSYYKCTYPN-CPTKKKVERSLDGQITEIV-YKGTHNHP 288
S D + WRKYGQK +KGS PR YY+C+ C +K+VER+ IV Y G HNHP
Sbjct: 76 SSDIWAWRKYGQKPIKGSPYPRGYYRCSSSKGCLARKQVERNKSEPAMFIVTYTGEHNHP 255
Query: 289 KPQATRRNSSSSSSLPIP 306
P T RNS + S+ P
Sbjct: 256 AP--THRNSLAGSTRQKP 303
>TC13725 similar to UP|Q39658 (Q39658) SPF1-like DNA-binding protein,
partial (11%)
Length = 578
Score = 72.0 bits (175), Expect = 2e-13
Identities = 39/79 (49%), Positives = 47/79 (59%), Gaps = 4/79 (5%)
Frame = +2
Query: 433 GCPVRKHVERASHDLRAVITTYEGKHNHDVPAARGSGNHSVT----RPLPNNTANAIRPS 488
GC VRKHVERAS D +AVITTYEGKHNHDVPAAR S +++ + +P P N P
Sbjct: 2 GCNVRKHVERASTDAKAVITTYEGKHNHDVPAARNSSHNTASSNSMQPKPQNMGPEKHPL 181
Query: 489 SVTHQHHHNTNNNSLQSLR 507
+N N+ LR
Sbjct: 182 LKDMDFGNNNNDQRPVHLR 238
Score = 32.3 bits (72), Expect = 0.22
Identities = 18/45 (40%), Positives = 26/45 (57%), Gaps = 3/45 (6%)
Frame = +2
Query: 262 CPTKKKVER-SLDGQITEIVYKGTHNHPKPQA--TRRNSSSSSSL 303
C +K VER S D + Y+G HNH P A + N++SS+S+
Sbjct: 5 CNVRKHVERASTDAKAVITTYEGKHNHDVPAARNSSHNTASSNSM 139
>TC8298 homologue to UP|Q9SXP4 (Q9SXP4) DNA-binding protein NtWRKY3,
partial (16%)
Length = 514
Score = 53.1 bits (126), Expect(2) = 3e-09
Identities = 24/47 (51%), Positives = 28/47 (59%), Gaps = 1/47 (2%)
Frame = +3
Query: 424 RSYYKCT-HPGCPVRKHVERASHDLRAVITTYEGKHNHDVPAARGSG 469
R YYKC+ GCP RKHVERA D +I TYEG+H H + G
Sbjct: 159 RGYYKCSSEKGCPARKHVERAQDDPNMLIVTYEGEHRHLITTTAAVG 299
Score = 25.0 bits (53), Expect(2) = 3e-09
Identities = 9/13 (69%), Positives = 11/13 (84%)
Frame = +1
Query: 412 YGQKVVKGNPNPR 424
YGQK +KG+P PR
Sbjct: 1 YGQKPIKGSPYPR 39
>TC16534 similar to UP|WR41_ARATH (Q8H0Y8) Probable WRKY transcription
factor 41 (WRKY DNA-binding protein 41), partial (20%)
Length = 697
Score = 56.6 bits (135), Expect = 1e-08
Identities = 21/30 (70%), Positives = 26/30 (86%)
Frame = +1
Query: 232 DDGYNWRKYGQKQVKGSENPRSYYKCTYPN 261
DDGYNWRKYGQK + G++ PRSYY+CT+ N
Sbjct: 586 DDGYNWRKYGQKDILGAKYPRSYYRCTFRN 675
Score = 48.5 bits (114), Expect = 3e-06
Identities = 19/27 (70%), Positives = 21/27 (77%)
Frame = +1
Query: 404 DDGYRWRKYGQKVVKGNPNPRSYYKCT 430
DDGY WRKYGQK + G PRSYY+CT
Sbjct: 586 DDGYNWRKYGQKDILGAKYPRSYYRCT 666
>TC11031 homologue to UP|WR65_ARATH (Q9LP56) Probable WRKY transcription
factor 65 (WRKY DNA-binding protein 65), partial (15%)
Length = 453
Score = 52.8 bits (125), Expect = 2e-07
Identities = 32/105 (30%), Positives = 51/105 (48%), Gaps = 14/105 (13%)
Frame = +3
Query: 347 EQSSQKCRSGGDELDEDEPDSKRWKVEG---ENEGISAPGSRTVREPRVV---VQTTSDI 400
E+ ++ + + + P S + ++G + SA SR + RVV ++ T
Sbjct: 138 EEDAEVAHHHQENIADSPPSSTAFNIDGLVTSSSTSSAKRSRRAIQKRVVQIPIKETEGS 317
Query: 401 DIL-------DDGYRWRKYGQKVVKGNPNPRSYYKC-THPGCPVR 437
+ D + WRKYGQK +KG+P PR YY+C + GCP R
Sbjct: 318 RLKAENNTPPSDSWAWRKYGQKPIKGSPYPRGYYRCSSSKGCPAR 452
Score = 45.8 bits (107), Expect = 2e-05
Identities = 19/39 (48%), Positives = 24/39 (60%), Gaps = 1/39 (2%)
Frame = +3
Query: 228 NRRSDDGYNWRKYGQKQVKGSENPRSYYKC-TYPNCPTK 265
N D + WRKYGQK +KGS PR YY+C + CP +
Sbjct: 336 NTPPSDSWAWRKYGQKPIKGSPYPRGYYRCSSSKGCPAR 452
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.309 0.125 0.366
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,330,032
Number of Sequences: 28460
Number of extensions: 192811
Number of successful extensions: 2244
Number of sequences better than 10.0: 174
Number of HSP's better than 10.0 without gapping: 1924
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2159
length of query: 579
length of database: 4,897,600
effective HSP length: 95
effective length of query: 484
effective length of database: 2,193,900
effective search space: 1061847600
effective search space used: 1061847600
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 58 (26.9 bits)
Lotus: description of TM0058a.3


