

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0590c.6
(364 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
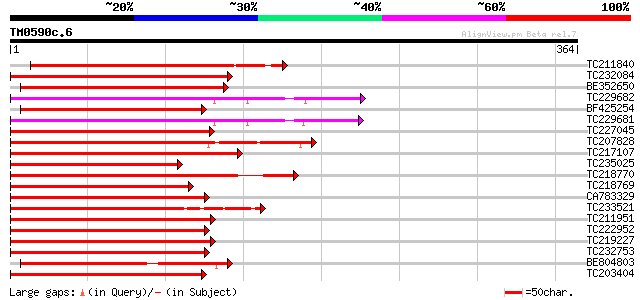
Score E
Sequences producing significant alignments: (bits) Value
TC211840 homologue to UP|Q8LBF0 (Q8LBF0) Myb-related protein M4,... 251 3e-67
TC232084 UP|Q84PP2 (Q84PP2) Transcription factor MYB101 (Fragmen... 228 4e-60
BE352650 221 4e-58
TC229682 UP|Q9XIU9 (Q9XIU9) GmMYB29A2 protein, complete 202 1e-52
BF425254 homologue to PIR|S26605|S26 myb-related protein 1 - gar... 202 1e-52
TC229681 UP|Q9S7E3 (Q9S7E3) GmMYB29A1 protein, complete 200 9e-52
TC227045 homologue to UP|Q43525 (Q43525) Transcription factor, p... 194 7e-50
TC207828 homologue to UP|Q6SL45 (Q6SL45) MYB6, partial (52%) 192 2e-49
TC217107 homologue to UP|Q7X9I1 (Q7X9I1) MYB transcription facto... 192 2e-49
TC235025 similar to UP|Q6R077 (Q6R077) MYB transcription factor,... 192 3e-49
TC218770 UP|Q9XIU5 (Q9XIU5) GmMYB29B2 protein, complete 188 4e-48
TC218769 UP|Q9XIU8 (Q9XIU8) GmMYB29A2 protein, complete 187 5e-48
CA783329 homologue to GP|6651292|gb| myb-related transcription f... 187 5e-48
TC233521 similar to UP|O04140 (O04140) Myb protein, partial (44%) 185 3e-47
TC211951 similar to GB|AAS10022.1|41619094|AY519552 MYB transcri... 184 5e-47
TC222952 homologue to UP|Q40173 (Q40173) Myb-related transcripti... 182 2e-46
TC219227 homologue to UP|Q84U52 (Q84U52) MYB2, partial (36%) 181 5e-46
TC232753 GmMYB10 178 4e-45
BE804803 homologue to GP|13346194|gb GHMYB9 {Gossypium hirsutum}... 177 5e-45
TC203404 GmMYB16 174 7e-44
>TC211840 homologue to UP|Q8LBF0 (Q8LBF0) Myb-related protein M4, partial
(38%)
Length = 516
Score = 251 bits (642), Expect = 3e-67
Identities = 120/166 (72%), Positives = 140/166 (84%), Gaps = 1/166 (0%)
Frame = +2
Query: 14 KGPWTPEEDQKLVDYIQKHGHGKWRTLPKNAGLKRCGKSCRLRWANYLRPDIKRGKFSLE 73
KGPWT EEDQKL+DYIQKHG+G WRTLPKNAGL+RCGKSCRLRW NYLRPDIKRG+FS E
Sbjct: 2 KGPWTTEEDQKLIDYIQKHGYGNWRTLPKNAGLQRCGKSCRLRWTNYLRPDIKRGRFSFE 181
Query: 74 EEEAIIQLHSVLGNKWSTIAANLQGRTDNEIKNYWNTHIKKKLLKMGLDPVTHTPRLDVL 133
EEE IIQLHS+LGNKWS IA+ L GRTDNEIKNYWNTHI+K+LL+MG+DPVTH+PRLD+L
Sbjct: 182 EEETIIQLHSILGNKWSAIASRLPGRTDNEIKNYWNTHIRKRLLRMGIDPVTHSPRLDLL 361
Query: 134 QLASILNSSLYNNSAQFNNPGLFGMER-VVNPSQLQLLNLVTTLLS 178
+ SILN+S Y S+Q N L GM + +VNP +LL L ++L S
Sbjct: 362 DINSILNASFY-GSSQINIQRLLGMHQYMVNP---ELLKLASSLFS 487
>TC232084 UP|Q84PP2 (Q84PP2) Transcription factor MYB101 (Fragment), complete
Length = 1383
Score = 228 bits (580), Expect = 4e-60
Identities = 105/143 (73%), Positives = 118/143 (82%)
Frame = +2
Query: 1 MAKSNSGEKNGVKKGPWTPEEDQKLVDYIQKHGHGKWRTLPKNAGLKRCGKSCRLRWANY 60
M ++ ++NG+KKGPWTPEED+ LVDYIQKHGHG WR LPK+A L RCGKSCRLRW NY
Sbjct: 53 MGRTPCSDENGLKKGPWTPEEDRILVDYIQKHGHGSWRALPKHARLNRCGKSCRLRWTNY 232
Query: 61 LRPDIKRGKFSLEEEEAIIQLHSVLGNKWSTIAANLQGRTDNEIKNYWNTHIKKKLLKMG 120
LRPDIKRGKFS EEE+ II LH+VLGNKWS IA +L GRTDNEIKN+WNTH+KKKLL+MG
Sbjct: 233 LRPDIKRGKFSEEEEQLIINLHAVLGNKWSAIAGHLPGRTDNEIKNFWNTHLKKKLLQMG 412
Query: 121 LDPVTHTPRLDVLQLASILNSSL 143
LDPVTH PR D L L S L L
Sbjct: 413 LDPVTHRPRSDHLNLLSNLQQLL 481
>BE352650
Length = 528
Score = 221 bits (563), Expect = 4e-58
Identities = 99/133 (74%), Positives = 111/133 (83%)
Frame = +1
Query: 8 EKNGVKKGPWTPEEDQKLVDYIQKHGHGKWRTLPKNAGLKRCGKSCRLRWANYLRPDIKR 67
E + VKKGPWTPEED+KL+DYI KHGHG WRTLPK AGL RCGKSCRLRW NYLRPDIKR
Sbjct: 127 ESSSVKKGPWTPEEDEKLIDYISKHGHGSWRTLPKRAGLNRCGKSCRLRWTNYLRPDIKR 306
Query: 68 GKFSLEEEEAIIQLHSVLGNKWSTIAANLQGRTDNEIKNYWNTHIKKKLLKMGLDPVTHT 127
GKFS ++E II HSVLGNKWS IAA+L GRTDNEIKNYW THI+KKLLKMG+DP T+
Sbjct: 307 GKFSEDDERIIINFHSVLGNKWSKIAAHLPGRTDNEIKNYWTTHIRKKLLKMGIDPETYK 486
Query: 128 PRLDVLQLASILN 140
PR D+ L S+ +
Sbjct: 487 PRTDLNHLMSLFS 525
>TC229682 UP|Q9XIU9 (Q9XIU9) GmMYB29A2 protein, complete
Length = 1249
Score = 202 bits (515), Expect = 1e-52
Identities = 104/239 (43%), Positives = 143/239 (59%), Gaps = 11/239 (4%)
Frame = +3
Query: 1 MAKSNSGEKNGVKKGPWTPEEDQKLVDYIQKHGHGKWRTLPKNAGLKRCGKSCRLRWANY 60
M ++ EK G+KKGPWTPEEDQ L+ YIQKHGHG WR LPK AGL RCGKSCRLRW NY
Sbjct: 84 MVRAPCCEKMGLKKGPWTPEEDQILMSYIQKHGHGNWRALPKLAGLLRCGKSCRLRWINY 263
Query: 61 LRPDIKRGKFSLEEEEAIIQLHSVLGNKWSTIAANLQGRTDNEIKNYWNTHIKKKLLKMG 120
LRPDIKRG FS EEEE II++H +LGN+WS IAA L GRTDNEIKN W+TH+KK+LL
Sbjct: 264 LRPDIKRGNFSSEEEEIIIKMHELLGNRWSAIAAKLPGRTDNEIKNVWHTHLKKRLLNSD 443
Query: 121 LDPVTHTPRL-------DVLQLASILNSSLYNNSAQFN--NPGLFGMERVVNPSQLQLLN 171
+ PR+ L +S+ + + F+ + G M+ ++ ++ +
Sbjct: 444 IQKRVSKPRIKRSDSNSSTLTQLEPTSSACTTSLSDFSSFSEGTKNMDNMIKSEDIESVE 623
Query: 172 LVTTLLSCQNTNQDVWN--TNNYQQNQLGGSTQWQHQTQCSQPMQLDNSIHAFQPNQVN 228
+ + ++ W+ T +Y+ + + S W + + P NS+ +FQ V+
Sbjct: 624 TIMPPI-----DESFWSEATVDYESSTMMTSNSWTISNELAPPQYQFNSVESFQQQSVD 785
>BF425254 homologue to PIR|S26605|S26 myb-related protein 1 - garden petunia,
partial (29%)
Length = 375
Score = 202 bits (515), Expect = 1e-52
Identities = 87/119 (73%), Positives = 104/119 (87%)
Frame = +3
Query: 8 EKNGVKKGPWTPEEDQKLVDYIQKHGHGKWRTLPKNAGLKRCGKSCRLRWANYLRPDIKR 67
+K G+KKGPWTPEEDQKL+ YI++HGHG WR LP AGL+RCGKSCRLRW NYLRPDIKR
Sbjct: 18 DKVGLKKGPWTPEEDQKLLAYIEEHGHGSWRALPAKAGLQRCGKSCRLRWTNYLRPDIKR 197
Query: 68 GKFSLEEEEAIIQLHSVLGNKWSTIAANLQGRTDNEIKNYWNTHIKKKLLKMGLDPVTH 126
GKF ++EE+ IIQLH++LGN+WS+I+ +L RTDNEIKNYWNTH+KK L KM +DPVTH
Sbjct: 198 GKFXMQEEQTIIQLHALLGNRWSSISTHLPKRTDNEIKNYWNTHLKKXLNKMSIDPVTH 374
>TC229681 UP|Q9S7E3 (Q9S7E3) GmMYB29A1 protein, complete
Length = 795
Score = 200 bits (508), Expect = 9e-52
Identities = 106/239 (44%), Positives = 141/239 (58%), Gaps = 12/239 (5%)
Frame = +1
Query: 1 MAKSNSGEKNGVKKGPWTPEEDQKLVDYIQKHGHGKWRTLPKNAGLKRCGKSCRLRWANY 60
M ++ EK G+KKGPWTPEEDQ L+ YIQKHGHG WR LPK AGL RCGKSCRLRW NY
Sbjct: 1 MVRAPCCEKMGLKKGPWTPEEDQILMSYIQKHGHGNWRALPKLAGLLRCGKSCRLRWINY 180
Query: 61 LRPDIKRGKFSLEEEEAIIQLHSVLGNKWSTIAANLQGRTDNEIKNYWNTHIKKKLLKMG 120
LRPDIKRG FS EEEE II++H +LGN+WS IAA L GRTDNEIKN W+TH+KK+L+
Sbjct: 181 LRPDIKRGNFSSEEEEIIIKMHELLGNRWSAIAAKLPGRTDNEIKNVWHTHLKKRLMNSD 360
Query: 121 LDPVTHTPRL-------DVLQLASILNSSLYNNSAQFN--NPGLFGMERVVNPSQLQLLN 171
+ PR+ L + +SS S+ F+ + G M+ ++ ++ +
Sbjct: 361 TNKRVSKPRIKRSDSNSSTLTQSEPTSSSGCTTSSDFSSFSEGTKNMDNMIKREDIESME 540
Query: 172 LVTTLLSCQNTNQDVW--NTNNYQQN-QLGGSTQWQHQTQCSQPMQLDNSIHAFQPNQV 227
V + ++ W T +Y+ + + S W + + P NS+ FQ V
Sbjct: 541 TVKPPI-----DESFWPQETVDYESSTMMQSSNSWTISNELAPPQYQFNSVETFQQQSV 702
>TC227045 homologue to UP|Q43525 (Q43525) Transcription factor, partial (55%)
Length = 560
Score = 194 bits (492), Expect = 7e-50
Identities = 88/131 (67%), Positives = 101/131 (76%)
Frame = +3
Query: 1 MAKSNSGEKNGVKKGPWTPEEDQKLVDYIQKHGHGKWRTLPKNAGLKRCGKSCRLRWANY 60
M +S EK KG WT EED +L+ YI+ HG G WR+LPK AGL RCGKSCRLRW NY
Sbjct: 15 MGRSPCCEKAHTNKGAWTKEEDHRLISYIRAHGEGCWRSLPKAAGLLRCGKSCRLRWINY 194
Query: 61 LRPDIKRGKFSLEEEEAIIQLHSVLGNKWSTIAANLQGRTDNEIKNYWNTHIKKKLLKMG 120
LRPD+KRG FSLEE++ II+LHS+LGNKWS IA L GRTDNEIKNYWNTHI++KLL G
Sbjct: 195 LRPDLKRGNFSLEEDQLIIKLHSLLGNKWSLIAGRLPGRTDNEIKNYWNTHIRRKLLSRG 374
Query: 121 LDPVTHTPRLD 131
+DP TH P D
Sbjct: 375 IDPATHRPLND 407
>TC207828 homologue to UP|Q6SL45 (Q6SL45) MYB6, partial (52%)
Length = 999
Score = 192 bits (489), Expect = 2e-49
Identities = 103/207 (49%), Positives = 131/207 (62%), Gaps = 10/207 (4%)
Frame = +2
Query: 1 MAKSNSGEKNGVKKGPWTPEEDQKLVDYIQKHGHGKWRTLPKNAGLKRCGKSCRLRWANY 60
M +S EK KG WT EED++L++YI+ HG G WR+LPK AGL RCGKSCRLRW NY
Sbjct: 41 MGRSPCCEKEHTNKGAWTKEEDERLINYIKLHGEGCWRSLPKAAGLLRCGKSCRLRWINY 220
Query: 61 LRPDIKRGKFSLEEEEAIIQLHSVLGNKWSTIAANLQGRTDNEIKNYWNTHIKKKLLKMG 120
LRPD+KRG F+ EE+E II LHS+LGNKWS IAA L GRTDNEIKNYWNTHIK+KL G
Sbjct: 221 LRPDLKRGNFTEEEDELIINLHSLLGNKWSLIAARLPGRTDNEIKNYWNTHIKRKLYSRG 400
Query: 121 LDPVTH--------TPRLDVLQLASILNSSLYNNSAQFNNPGLFGMERVVNPSQLQLLNL 172
+DP TH TP V A+ + SS + NN G + +VN S +
Sbjct: 401 IDPQTHRPLNAASATPATTV--TATAVPSSNSKKKIKNNNNNNNGFQ-LVNNSAYANTKI 571
Query: 173 VTTLLSCQNTNQD--VWNTNNYQQNQL 197
T +++ +++N V ++ +QL
Sbjct: 572 GTDMIAAEDSNSSSGVTTEESFPHHQL 652
>TC217107 homologue to UP|Q7X9I1 (Q7X9I1) MYB transcription factor R2R3 type,
partial (68%)
Length = 1058
Score = 192 bits (489), Expect = 2e-49
Identities = 88/149 (59%), Positives = 106/149 (71%)
Frame = +2
Query: 1 MAKSNSGEKNGVKKGPWTPEEDQKLVDYIQKHGHGKWRTLPKNAGLKRCGKSCRLRWANY 60
M +S EK KG WT EED +L+ YI+ HG G WR+LPK AGL RCGKSCRLRW NY
Sbjct: 266 MGRSPCCEKAHTNKGAWTKEEDDRLISYIRAHGEGCWRSLPKAAGLLRCGKSCRLRWINY 445
Query: 61 LRPDIKRGKFSLEEEEAIIQLHSVLGNKWSTIAANLQGRTDNEIKNYWNTHIKKKLLKMG 120
LRPD+KRG F+ EE+E II+LHS+LGNKWS IA L GRTDNEIKNYWNTHI++KLL G
Sbjct: 446 LRPDLKRGNFTEEEDELIIKLHSLLGNKWSLIAGRLPGRTDNEIKNYWNTHIRRKLLNRG 625
Query: 121 LDPVTHTPRLDVLQLASILNSSLYNNSAQ 149
+DP TH P + A++ + + Q
Sbjct: 626 IDPATHRPLNEAATAATVTTNISFGKQEQ 712
>TC235025 similar to UP|Q6R077 (Q6R077) MYB transcription factor, partial
(35%)
Length = 434
Score = 192 bits (487), Expect = 3e-49
Identities = 88/112 (78%), Positives = 94/112 (83%), Gaps = 1/112 (0%)
Frame = +1
Query: 1 MAKSNSGEKN-GVKKGPWTPEEDQKLVDYIQKHGHGKWRTLPKNAGLKRCGKSCRLRWAN 59
M +S E+N GVKKGPWTPEED+KL+DYI KHGHG WRTLPK AGL RCGKSCRLRW N
Sbjct: 97 MGRSPCCEENVGVKKGPWTPEEDEKLIDYISKHGHGSWRTLPKRAGLNRCGKSCRLRWTN 276
Query: 60 YLRPDIKRGKFSLEEEEAIIQLHSVLGNKWSTIAANLQGRTDNEIKNYWNTH 111
YL PDIKRGKFS E+E II LHSVLGNKWS IA +L GRTDNEIKNYWNTH
Sbjct: 277 YLTPDIKRGKFSEEDERIIINLHSVLGNKWSKIATHLPGRTDNEIKNYWNTH 432
>TC218770 UP|Q9XIU5 (Q9XIU5) GmMYB29B2 protein, complete
Length = 1316
Score = 188 bits (477), Expect = 4e-48
Identities = 95/185 (51%), Positives = 115/185 (61%)
Frame = +1
Query: 1 MAKSNSGEKNGVKKGPWTPEEDQKLVDYIQKHGHGKWRTLPKNAGLKRCGKSCRLRWANY 60
M ++ EK G+KKGPW EEDQ L YIQKHGHG WR LPK AGL RCGKSCRLRW NY
Sbjct: 112 MVRAPCCEKMGLKKGPWATEEDQILTSYIQKHGHGNWRALPKQAGLLRCGKSCRLRWINY 291
Query: 61 LRPDIKRGKFSLEEEEAIIQLHSVLGNKWSTIAANLQGRTDNEIKNYWNTHIKKKLLKMG 120
LRPDIKRG F++EEEE II+LH +LGN+WS IAA L GRTDNEIKN W+T++KK+LLK
Sbjct: 292 LRPDIKRGNFTIEEEETIIKLHEMLGNRWSAIAAKLPGRTDNEIKNVWHTNLKKRLLKSD 471
Query: 121 LDPVTHTPRLDVLQLASILNSSLYNNSAQFNNPGLFGMERVVNPSQLQLLNLVTTLLSCQ 180
+ S NSS+ S P+ L+ + TT +C
Sbjct: 472 QSKPSSKRATKPKIKRSDSNSSIITQS---------------EPAHLRFREMDTTSTACN 606
Query: 181 NTNQD 185
++ D
Sbjct: 607 TSSSD 621
>TC218769 UP|Q9XIU8 (Q9XIU8) GmMYB29A2 protein, complete
Length = 828
Score = 187 bits (476), Expect = 5e-48
Identities = 84/118 (71%), Positives = 96/118 (81%)
Frame = +1
Query: 1 MAKSNSGEKNGVKKGPWTPEEDQKLVDYIQKHGHGKWRTLPKNAGLKRCGKSCRLRWANY 60
M ++ EK G+KKGPW PEEDQ L YI KHGHG WR LPK AGL RCGKSCRLRW NY
Sbjct: 1 MVRAPCCEKMGLKKGPWAPEEDQILTSYIDKHGHGNWRALPKQAGLLRCGKSCRLRWINY 180
Query: 61 LRPDIKRGKFSLEEEEAIIQLHSVLGNKWSTIAANLQGRTDNEIKNYWNTHIKKKLLK 118
LRPDIKRG F++EEEE II+LH +LGN+WS IAA L GRTDNEIKN W+T++KK+LLK
Sbjct: 181 LRPDIKRGNFTIEEEETIIKLHDMLGNRWSAIAAKLPGRTDNEIKNVWHTNLKKRLLK 354
>CA783329 homologue to GP|6651292|gb| myb-related transcription factor
{Pimpinella brachycarpa}, partial (48%)
Length = 443
Score = 187 bits (476), Expect = 5e-48
Identities = 85/128 (66%), Positives = 102/128 (79%)
Frame = +3
Query: 1 MAKSNSGEKNGVKKGPWTPEEDQKLVDYIQKHGHGKWRTLPKNAGLKRCGKSCRLRWANY 60
M + +K GVKKGPWT EED+KL+++I +G WR +PK AGLKRCGKSCRLRW NY
Sbjct: 3 MGRQPRCDKLGVKKGPWTAEEDKKLINFILTNGQCCWRAVPKLAGLKRCGKSCRLRWTNY 182
Query: 61 LRPDIKRGKFSLEEEEAIIQLHSVLGNKWSTIAANLQGRTDNEIKNYWNTHIKKKLLKMG 120
LRPD+KRG + EE+ +I LH+ LGN+WS IAA L GRTDNEIKN+WNTHIKKKLLK+G
Sbjct: 183 LRPDLKRGLLTEAEEQLVIDLHARLGNRWSKIAARLPGRTDNEIKNHWNTHIKKKLLKIG 362
Query: 121 LDPVTHTP 128
+DPVTH P
Sbjct: 363 IDPVTHEP 386
>TC233521 similar to UP|O04140 (O04140) Myb protein, partial (44%)
Length = 886
Score = 185 bits (469), Expect = 3e-47
Identities = 94/164 (57%), Positives = 117/164 (71%)
Frame = +2
Query: 1 MAKSNSGEKNGVKKGPWTPEEDQKLVDYIQKHGHGKWRTLPKNAGLKRCGKSCRLRWANY 60
M ++ E+ G+KKGPWT EEDQ LV +IQ++GHG WR LPK AGL RCGKSCRLRW NY
Sbjct: 20 MTRTPCCERMGLKKGPWTAEEDQILVSHIQRYGHGNWRALPKQAGLLRCGKSCRLRWINY 199
Query: 61 LRPDIKRGKFSLEEEEAIIQLHSVLGNKWSTIAANLQGRTDNEIKNYWNTHIKKKLLKMG 120
LRPDIKRGKFS EEE+ I++LH +LGN+WS IAA+L GRTDNEIKN+W+TH+ K++ K G
Sbjct: 200 LRPDIKRGKFSKEEEDTILKLHGILGNRWSAIAASLPGRTDNEIKNFWHTHL-KRIQKSG 376
Query: 121 LDPVTHTPRLDVLQLASILNSSLYNNSAQFNNPGLFGMERVVNP 164
+ P +LQ A N+S +S N GL R +NP
Sbjct: 377 VH--NGNPSSRILQEAQ-ANTSSNASSVMIANYGL--PSRNINP 493
>TC211951 similar to GB|AAS10022.1|41619094|AY519552 MYB transcription factor
{Arabidopsis thaliana;} , partial (38%)
Length = 776
Score = 184 bits (467), Expect = 5e-47
Identities = 82/132 (62%), Positives = 105/132 (79%)
Frame = +2
Query: 1 MAKSNSGEKNGVKKGPWTPEEDQKLVDYIQKHGHGKWRTLPKNAGLKRCGKSCRLRWANY 60
M + + K ++KG W+PEED+KL++YI KHGHG W ++PK AGL+RCGKSCRLRW NY
Sbjct: 92 MGRHSCCYKQKLRKGLWSPEEDEKLLNYITKHGHGCWSSVPKLAGLQRCGKSCRLRWINY 271
Query: 61 LRPDIKRGKFSLEEEEAIIQLHSVLGNKWSTIAANLQGRTDNEIKNYWNTHIKKKLLKMG 120
LRPD+KRG FS +EE +II+LH+VLGN+WS IAA L GRTDNEIKN WN+ +KKKL + G
Sbjct: 272 LRPDLKRGAFSQQEENSIIELHAVLGNRWSQIAAQLPGRTDNEIKNLWNSCLKKKLRQRG 451
Query: 121 LDPVTHTPRLDV 132
+DP TH P ++
Sbjct: 452 IDPNTHQPLSEI 487
>TC222952 homologue to UP|Q40173 (Q40173) Myb-related transcription factor,
partial (47%)
Length = 612
Score = 182 bits (463), Expect = 2e-46
Identities = 83/128 (64%), Positives = 100/128 (77%)
Frame = +2
Query: 1 MAKSNSGEKNGVKKGPWTPEEDQKLVDYIQKHGHGKWRTLPKNAGLKRCGKSCRLRWANY 60
M + +K G+KKGPWT EED+KL+ +I +G WR +PK AGL RCGKSCRLRW NY
Sbjct: 125 MGRQPCCDKVGLKKGPWTAEEDKKLISFILTNGQCCWRAVPKLAGLLRCGKSCRLRWTNY 304
Query: 61 LRPDIKRGKFSLEEEEAIIQLHSVLGNKWSTIAANLQGRTDNEIKNYWNTHIKKKLLKMG 120
LRPD+KRG S EE+ +I LH+ LGN+WS IA++L GRTDNEIKN+WNTHIKKKL KMG
Sbjct: 305 LRPDLKRGLLSEYEEKMVIDLHAQLGNRWSKIASHLPGRTDNEIKNHWNTHIKKKLKKMG 484
Query: 121 LDPVTHTP 128
+DPVTH P
Sbjct: 485 IDPVTHKP 508
>TC219227 homologue to UP|Q84U52 (Q84U52) MYB2, partial (36%)
Length = 1311
Score = 181 bits (459), Expect = 5e-46
Identities = 82/132 (62%), Positives = 103/132 (77%)
Frame = +1
Query: 1 MAKSNSGEKNGVKKGPWTPEEDQKLVDYIQKHGHGKWRTLPKNAGLKRCGKSCRLRWANY 60
M + + K ++KG W+PEED+KL+ +I K+GHG W ++PK AGL+RCGKSCRLRW NY
Sbjct: 4 MGRHSCCYKQKLRKGLWSPEEDEKLLRHITKYGHGCWSSVPKQAGLQRCGKSCRLRWINY 183
Query: 61 LRPDIKRGKFSLEEEEAIIQLHSVLGNKWSTIAANLQGRTDNEIKNYWNTHIKKKLLKMG 120
LRPD+ RG FS EEE II+LH+VLGN+WS IAA L GRTDNEIKN WN+ +KKKL + G
Sbjct: 184 LRPDLNRGTFSQEEETLIIELHAVLGNRWSQIAAQLPGRTDNEIKNLWNSCLKKKLRQRG 363
Query: 121 LDPVTHTPRLDV 132
+DPVTH P +V
Sbjct: 364 IDPVTHKPLSEV 399
>TC232753 GmMYB10
Length = 423
Score = 178 bits (451), Expect = 4e-45
Identities = 80/128 (62%), Positives = 100/128 (77%)
Frame = +2
Query: 1 MAKSNSGEKNGVKKGPWTPEEDQKLVDYIQKHGHGKWRTLPKNAGLKRCGKSCRLRWANY 60
M + + K ++KG W+PEED+KL +YI + G G W ++PK AGL+RCGKSCRLRW NY
Sbjct: 32 MGRHSCCVKQKLRKGLWSPEEDEKLFNYITRFGVGCWSSVPKLAGLQRCGKSCRLRWINY 211
Query: 61 LRPDIKRGKFSLEEEEAIIQLHSVLGNKWSTIAANLQGRTDNEIKNYWNTHIKKKLLKMG 120
LRPD+KRG FS +EE+ II LH VLGN+W+ IAA L GRTDNEIKN+WN+ +KKKLLK G
Sbjct: 212 LRPDLKRGMFSQQEEDLIISLHEVLGNRWAQIAAQLPGRTDNEIKNFWNSCLKKKLLKQG 391
Query: 121 LDPVTHTP 128
+DP TH P
Sbjct: 392 IDPSTHKP 415
>BE804803 homologue to GP|13346194|gb GHMYB9 {Gossypium hirsutum}, partial
(50%)
Length = 421
Score = 177 bits (450), Expect = 5e-45
Identities = 87/138 (63%), Positives = 100/138 (72%), Gaps = 2/138 (1%)
Frame = +2
Query: 8 EKNGVKKGPWTPEEDQKLVDYIQKHGHGKWRTLPKNAGLKRCGKSCRLRWANYLRPDIKR 67
EK KG WT EED +L+ YI+ HG G WR+LPK AGL RCGKSCRLRW NYLRPD+KR
Sbjct: 5 EKAHTNKGAWTKEEDHRLISYIRAHGEGCWRSLPKAAGLLRCGKSCRLRWINYLRPDLKR 184
Query: 68 GKFSLEEEEAIIQLHSVLGNKWSTIAANLQGRTDNEIKNYWNTHIKKKLLKMGLDPVTHT 127
G FSLEE++ II+LHS+LGNK L GRTDNEIKNYWNTHI++KLL G+DP TH
Sbjct: 185 GNFSLEEDQLIIKLHSLLGNK-------LPGRTDNEIKNYWNTHIRRKLLSRGIDPATHR 343
Query: 128 PRLD--VLQLASILNSSL 143
P D VL+ LN L
Sbjct: 344 PLNDDKVLERCPDLNLEL 397
>TC203404 GmMYB16
Length = 1138
Score = 174 bits (440), Expect = 7e-44
Identities = 80/127 (62%), Positives = 95/127 (73%), Gaps = 1/127 (0%)
Frame = +1
Query: 1 MAKSNSGEKNGVKKGPWTPEEDQKLVDYIQKHGHGKWRTLPKNAGLKRCGKSCRLRWANY 60
M ++ K G+ KGPWTP+ED L YIQ HG G+W++LPK AGL RCGKSCRLRW NY
Sbjct: 106 MGRAPCCSKVGLHKGPWTPKEDALLTKYIQAHGEGQWKSLPKKAGLLRCGKSCRLRWMNY 285
Query: 61 LRPDIKRGKFSLEEEEAIIQLHSVLGNKWSTIAANLQGRTDNEIKNYWNTHIKKKLLKMG 120
LRPDIKRG + EE++ II++HS+LGN+WS IA L GRTDNEIKNYWNTH+ KKL G
Sbjct: 286 LRPDIKRGNIAPEEDDLIIRMHSLLGNRWSLIAGRLPGRTDNEIKNYWNTHLSKKLKIQG 465
Query: 121 L-DPVTH 126
D TH
Sbjct: 466 TEDTDTH 486
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.311 0.128 0.381
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 19,485,793
Number of Sequences: 63676
Number of extensions: 342448
Number of successful extensions: 3234
Number of sequences better than 10.0: 288
Number of HSP's better than 10.0 without gapping: 2634
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2957
length of query: 364
length of database: 12,639,632
effective HSP length: 99
effective length of query: 265
effective length of database: 6,335,708
effective search space: 1678962620
effective search space used: 1678962620
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0590c.6


