

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
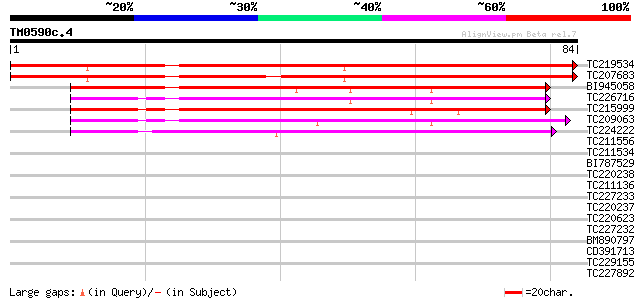
Query= TM0590c.4
(84 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC219534 similar to UP|Q04129 (Q04129) Wound induced protein (Fr... 83 3e-17
TC207683 similar to GB|AAO42915.1|28416769|BT004669 At4g10262 {A... 82 4e-17
BI945058 54 1e-08
TC226716 weakly similar to UP|O82615 (O82615) T9A4.6 protein (Pr... 46 3e-06
TC215999 similar to UP|O82615 (O82615) T9A4.6 protein (Probable ... 44 1e-05
TC209063 similar to UP|Q04129 (Q04129) Wound induced protein (Fr... 43 2e-05
TC224222 similar to UP|O82615 (O82615) T9A4.6 protein (Probable ... 39 6e-04
TC211556 similar to UP|Q04129 (Q04129) Wound induced protein (Fr... 36 0.003
TC211534 homologue to GB|AAO42915.1|28416769|BT004669 At4g10262 ... 33 0.033
BI787529 homologue to GP|29423283|gb envelope-like protein {Glyc... 28 0.62
TC220238 UP|Q84VH5 (Q84VH5) Envelope-like protein (Fragment), co... 28 0.62
TC211136 UP|Q84VI3 (Q84VI3) Envelope-like protein (Fragment), co... 28 1.1
TC227233 similar to GB|AAO42915.1|28416769|BT004669 At4g10262 {A... 27 2.4
TC220237 UP|O64406 (O64406) Envelope-like (Fragment), complete 27 2.4
TC220623 similar to UP|Q94AB4 (Q94AB4) AT3g13340/MDC11_13, parti... 27 2.4
TC227232 homologue to UP|Q04129 (Q04129) Wound induced protein (... 26 3.1
BM890797 26 3.1
CD391713 25 5.3
TC229155 similar to UP|Q75GV2 (Q75GV2) Expressed protein, partia... 25 6.9
TC227892 similar to UP|Q94AB4 (Q94AB4) AT3g13340/MDC11_13, parti... 25 9.0
>TC219534 similar to UP|Q04129 (Q04129) Wound induced protein (Fragment),
partial (84%)
Length = 492
Score = 82.8 bits (203), Expect = 3e-17
Identities = 49/91 (53%), Positives = 65/91 (70%), Gaps = 7/91 (7%)
Frame = +1
Query: 1 MSAATRSLIV----GAVEALKDKLGVYNRRRNYALRSLQQRVVKNNNITPNG---NSSSA 53
MSAA+R+ IV GAVEALKD+LGV R N+ALRSLQQ N +S+++
Sbjct: 49 MSAASRAWIVASSIGAVEALKDQLGVC--RWNHALRSLQQHAKSNIRSYTQAKTLSSATS 222
Query: 54 AAIPSKVKRSNQEFMRKVMDLNCWGPSTTKF 84
AA+ +KVKR+ +E MRKVMDL+CWGP+T++F
Sbjct: 223 AAVSNKVKRTKEESMRKVMDLSCWGPNTSRF 315
>TC207683 similar to GB|AAO42915.1|28416769|BT004669 At4g10262 {Arabidopsis
thaliana;} , partial (69%)
Length = 741
Score = 82.4 bits (202), Expect = 4e-17
Identities = 51/93 (54%), Positives = 64/93 (67%), Gaps = 9/93 (9%)
Frame = +3
Query: 1 MSAATRSLIV----GAVEALKDKLGVYNRRRNYALRSLQQRVVKNNNITPNG-----NSS 51
MSA+T++ IV GAVEALKD+LGV R NYALRSLQQ NNI +S+
Sbjct: 81 MSASTKAWIVASSIGAVEALKDQLGVC--RWNYALRSLQQHA--KNNIRSYSQARKLSSA 248
Query: 52 SAAAIPSKVKRSNQEFMRKVMDLNCWGPSTTKF 84
S+AA+ +KVKR+ +E M KV++ NCWGPST KF
Sbjct: 249 SSAAVSNKVKRTKEEHMGKVIEFNCWGPSTAKF 347
>BI945058
Length = 401
Score = 53.9 bits (128), Expect = 1e-08
Identities = 36/78 (46%), Positives = 50/78 (63%), Gaps = 7/78 (8%)
Frame = +1
Query: 10 VGAVEALKDKLGVYNRRRNYALRSLQQRVVKN-NNITPNGN-SSSAAAIPSKVK-----R 62
VGAVEALKD+LGV R NY LR QQ + + +++ N SSS+A + SK+K +
Sbjct: 46 VGAVEALKDQLGVC--RWNYVLRCAQQHMKNHFRSLSQAKNVSSSSALVASKLKGDEKAK 219
Query: 63 SNQEFMRKVMDLNCWGPS 80
+E +R VM L+CWGP+
Sbjct: 220 KAEESLRTVMYLSCWGPN 273
>TC226716 weakly similar to UP|O82615 (O82615) T9A4.6 protein (Probable
wound-induced protein), partial (78%)
Length = 658
Score = 46.2 bits (108), Expect = 3e-06
Identities = 33/79 (41%), Positives = 47/79 (58%), Gaps = 8/79 (10%)
Frame = +3
Query: 10 VGAVEALKDKLGVYNRRRNYALRSLQQRVVKNNNITPNGN----SSSAAAIPSKVK---- 61
VG VEALKD+ G+ R N ALRS Q +V + N SSS+A + S++K
Sbjct: 276 VGVVEALKDQ-GLC--RWNNALRSAQYQVKNHVRSLSQANKVSSSSSSAVVSSRLKEEGA 446
Query: 62 RSNQEFMRKVMDLNCWGPS 80
+ ++E +R VM L+CWGP+
Sbjct: 447 KQSEESLRTVMYLSCWGPN 503
>TC215999 similar to UP|O82615 (O82615) T9A4.6 protein (Probable
wound-induced protein), partial (50%)
Length = 773
Score = 44.3 bits (103), Expect = 1e-05
Identities = 32/76 (42%), Positives = 46/76 (60%), Gaps = 5/76 (6%)
Frame = +2
Query: 10 VGAVEALKDKLGVYNRRRNYALRSLQQRVVKNNNITPNGNSSSAAAIPS---KVKRSNQ- 65
VG VEALKD+ G+ R N+ALRS Q + + N S++A+ S K +++NQ
Sbjct: 170 VGIVEALKDQ-GIC--RWNHALRSAQHHLKNHVGSFSQANKLSSSAMISTTLKHEKANQS 340
Query: 66 -EFMRKVMDLNCWGPS 80
E +R VM L+CWGP+
Sbjct: 341 EESLRTVMYLSCWGPN 388
>TC209063 similar to UP|Q04129 (Q04129) Wound induced protein (Fragment),
partial (53%)
Length = 739
Score = 43.1 bits (100), Expect = 2e-05
Identities = 32/81 (39%), Positives = 45/81 (55%), Gaps = 7/81 (8%)
Frame = +2
Query: 10 VGAVEALKDKLGVYNRRRNYALRSLQQRVVKNNNI---TPNGNSSSAAAIPSKVK----R 62
VG VEALKD+ G+ R N +RS QQ N T + S+A +K+K +
Sbjct: 119 VGVVEALKDQ-GIC--RWNSVMRSAQQHAKHNMRSLSQTKKFSFQSSAMASAKLKDEKAK 289
Query: 63 SNQEFMRKVMDLNCWGPSTTK 83
++E +R VM L+CWGP+ TK
Sbjct: 290 QSEESLRTVMYLSCWGPN*TK 352
>TC224222 similar to UP|O82615 (O82615) T9A4.6 protein (Probable
wound-induced protein), partial (40%)
Length = 466
Score = 38.5 bits (88), Expect = 6e-04
Identities = 25/79 (31%), Positives = 42/79 (52%), Gaps = 7/79 (8%)
Frame = +1
Query: 10 VGAVEALKDKLGVYNRRRNYALRSLQQRV-------VKNNNITPNGNSSSAAAIPSKVKR 62
VG VEALKD+ Y + N ++S+ Q+ + + SSS+A ++ +R
Sbjct: 121 VGVVEALKDQ--GYCKMNNTMMKSVAQQAKSHLGSATRAKKLASPSPSSSSATSNNEKRR 294
Query: 63 SNQEFMRKVMDLNCWGPST 81
+E +R VM L+ WGP++
Sbjct: 295 MAEESLRTVMYLSTWGPNS 351
>TC211556 similar to UP|Q04129 (Q04129) Wound induced protein (Fragment),
partial (49%)
Length = 388
Score = 36.2 bits (82), Expect = 0.003
Identities = 34/89 (38%), Positives = 51/89 (57%), Gaps = 9/89 (10%)
Frame = +2
Query: 1 MSAATRSLIV----GAVEALKDKLGVYNRRRNYALRSLQQRV---VKNNNITPNGN-SSS 52
MS++ R+ IV G VEALKD+ GV+ N+ L+S Q + V + + N + SSS
Sbjct: 131 MSSSQRAWIVAASVGVVEALKDQ-GVFFC--NHTLKSAQHVINCHVGSFSQAKNLSFSSS 301
Query: 53 AAAIPSKVKRSNQE-FMRKVMDLNCWGPS 80
+ S++K E +R VM L+CWGP+
Sbjct: 302 MGSTSSRLKGEESEGALRTVMYLSCWGPN 388
>TC211534 homologue to GB|AAO42915.1|28416769|BT004669 At4g10262 {Arabidopsis
thaliana;} , partial (25%)
Length = 274
Score = 32.7 bits (73), Expect = 0.033
Identities = 18/68 (26%), Positives = 35/68 (51%)
Frame = +2
Query: 13 VEALKDKLGVYNRRRNYALRSLQQRVVKNNNITPNGNSSSAAAIPSKVKRSNQEFMRKVM 72
++ +K++LG +L+ K N +G S +K+K++ ++ +R VM
Sbjct: 56 LQGMKEQLGSNKCDSTIKSLNLRDSSSKQNRFLSSGTSFKPPNNDNKLKQA-EDSLRTVM 232
Query: 73 DLNCWGPS 80
L+CWGP+
Sbjct: 233 YLSCWGPN 256
>BI787529 homologue to GP|29423283|gb envelope-like protein {Glycine max},
partial (21%)
Length = 455
Score = 28.5 bits (62), Expect = 0.62
Identities = 17/55 (30%), Positives = 30/55 (53%)
Frame = +3
Query: 8 LIVGAVEALKDKLGVYNRRRNYALRSLQQRVVKNNNITPNGNSSSAAAIPSKVKR 62
L G E L+ + G +R+ ++++ Q+ K+ ITP + S AIPSK ++
Sbjct: 270 LAPGIAERLQSRKGKTPIKRSGRIKTMAQK--KSTPITPTTSRRSKVAIPSKKRK 428
>TC220238 UP|Q84VH5 (Q84VH5) Envelope-like protein (Fragment), complete
Length = 2128
Score = 28.5 bits (62), Expect = 0.62
Identities = 17/55 (30%), Positives = 30/55 (53%)
Frame = +1
Query: 8 LIVGAVEALKDKLGVYNRRRNYALRSLQQRVVKNNNITPNGNSSSAAAIPSKVKR 62
L G E L+ + G +R+ ++++ Q+ K+ ITP + S AIPSK ++
Sbjct: 742 LAPGIAERLQSRKGKTPIKRSGRIKTMAQK--KSTPITPTTSRRSKVAIPSKKRK 900
>TC211136 UP|Q84VI3 (Q84VI3) Envelope-like protein (Fragment), complete
Length = 2043
Score = 27.7 bits (60), Expect = 1.1
Identities = 17/55 (30%), Positives = 30/55 (53%)
Frame = +1
Query: 8 LIVGAVEALKDKLGVYNRRRNYALRSLQQRVVKNNNITPNGNSSSAAAIPSKVKR 62
L G E L+ + G +R+ ++++ Q+ K+ ITP + S AIPSK ++
Sbjct: 718 LAPGIAERLQSRKGKTPIKRSGRIKTMAQK--KSTPITPATSRRSKVAIPSKKRK 876
>TC227233 similar to GB|AAO42915.1|28416769|BT004669 At4g10262 {Arabidopsis
thaliana;} , partial (19%)
Length = 571
Score = 26.6 bits (57), Expect = 2.4
Identities = 10/31 (32%), Positives = 23/31 (73%)
Frame = +2
Query: 48 GNSSSAAAIPSKVKRSNQEFMRKVMDLNCWG 78
G+ + ++ + ++V++++ +RKVM L+CWG
Sbjct: 215 GSVAGSSDVENRVRQADDS-LRKVMYLSCWG 304
>TC220237 UP|O64406 (O64406) Envelope-like (Fragment), complete
Length = 1974
Score = 26.6 bits (57), Expect = 2.4
Identities = 17/55 (30%), Positives = 29/55 (51%)
Frame = +1
Query: 8 LIVGAVEALKDKLGVYNRRRNYALRSLQQRVVKNNNITPNGNSSSAAAIPSKVKR 62
L G E L+ + G R+ ++++ Q+ K+ ITP + S AIPSK ++
Sbjct: 679 LAPGIAERLQSRKGKTPITRSGRIKTMAQK--KSTPITPTTSRWSKVAIPSKKRK 837
>TC220623 similar to UP|Q94AB4 (Q94AB4) AT3g13340/MDC11_13, partial (31%)
Length = 562
Score = 26.6 bits (57), Expect = 2.4
Identities = 14/28 (50%), Positives = 15/28 (53%)
Frame = +3
Query: 2 SAATRSLIVGAVEALKDKLGVYNRRRNY 29
S T SL VG + L YNRRRNY
Sbjct: 321 SPDTESLFVGVWDRTYGSLLQYNRRRNY 404
>TC227232 homologue to UP|Q04129 (Q04129) Wound induced protein (Fragment),
partial (16%)
Length = 852
Score = 26.2 bits (56), Expect = 3.1
Identities = 15/57 (26%), Positives = 31/57 (54%), Gaps = 1/57 (1%)
Frame = +3
Query: 23 YNRRRNYALRSLQQRVVKNNNITPNGNSSSAAAIPSKVKRSN-QEFMRKVMDLNCWG 78
+NR R ++ +L + + P+ + ++A + +K + S + +RKVM +CWG
Sbjct: 183 HNRSRLFSGGALSDLRPLSGVVGPDVSGAAAGSSDAKNRVSQADDSLRKVMYFSCWG 353
>BM890797
Length = 421
Score = 26.2 bits (56), Expect = 3.1
Identities = 15/44 (34%), Positives = 30/44 (68%), Gaps = 1/44 (2%)
Frame = +1
Query: 2 SAATRSLIVGAVEALKDKLGVYN-RRRNYALRSLQQRVVKNNNI 44
++ TRS ++G V+ + ++GV + RRR+Y+ +S+ R V+N +
Sbjct: 286 NSRTRSDVLGTVDEKRLQIGVSSRRRRDYSCKSI--RGVENEQV 411
>CD391713
Length = 594
Score = 25.4 bits (54), Expect = 5.3
Identities = 9/16 (56%), Positives = 12/16 (74%)
Frame = -3
Query: 65 QEFMRKVMDLNCWGPS 80
+E R VM L+CWGP+
Sbjct: 511 EESFRTVMYLSCWGPN 464
>TC229155 similar to UP|Q75GV2 (Q75GV2) Expressed protein, partial (65%)
Length = 1143
Score = 25.0 bits (53), Expect = 6.9
Identities = 16/41 (39%), Positives = 21/41 (51%), Gaps = 10/41 (24%)
Frame = +2
Query: 36 QRVVKNNNI----------TPNGNSSSAAAIPSKVKRSNQE 66
QR++K N T G SSSAAA+ S K+ N+E
Sbjct: 497 QRLIKVTNFCLESQSQSMDTQKGPSSSAAAVTSCRKKKNEE 619
>TC227892 similar to UP|Q94AB4 (Q94AB4) AT3g13340/MDC11_13, partial (68%)
Length = 1179
Score = 24.6 bits (52), Expect = 9.0
Identities = 12/28 (42%), Positives = 15/28 (52%)
Frame = +3
Query: 2 SAATRSLIVGAVEALKDKLGVYNRRRNY 29
S T SL +G + L +NRRRNY
Sbjct: 816 SPDTESLFIGVWDRTYGSLLQFNRRRNY 899
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.315 0.126 0.352
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,108,422
Number of Sequences: 63676
Number of extensions: 31617
Number of successful extensions: 220
Number of sequences better than 10.0: 42
Number of HSP's better than 10.0 without gapping: 209
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 215
length of query: 84
length of database: 12,639,632
effective HSP length: 60
effective length of query: 24
effective length of database: 8,819,072
effective search space: 211657728
effective search space used: 211657728
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 52 (24.6 bits)
Lotus: description of TM0590c.4


