

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0371a.1
(409 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
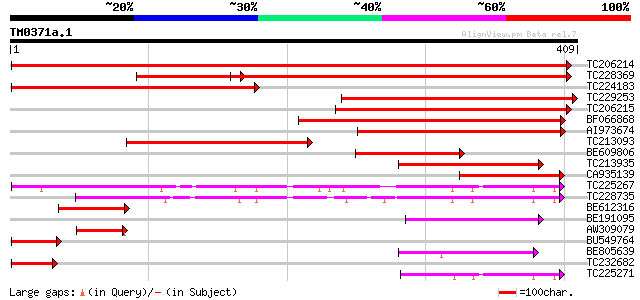
Score E
Sequences producing significant alignments: (bits) Value
TC206214 beta-D-glucan exohydrolase [Glycine max] 606 e-174
TC228369 similar to UP|Q7XAS3 (Q7XAS3) Beta-D-glucosidase, parti... 374 e-104
TC224183 similar to UP|Q7XAS3 (Q7XAS3) Beta-D-glucosidase, parti... 324 5e-89
TC229253 similar to UP|Q7XAS3 (Q7XAS3) Beta-D-glucosidase, parti... 311 3e-85
TC206215 similar to UP|Q7XAS3 (Q7XAS3) Beta-D-glucosidase, parti... 277 7e-75
BF066868 weakly similar to PIR|T51283|T51 glucan 1 3-beta-glucos... 258 3e-69
AI973674 weakly similar to PIR|T10521|T10 beta-glucosidase (EC 3... 198 3e-51
TC213093 similar to UP|Q7XAS3 (Q7XAS3) Beta-D-glucosidase, parti... 191 4e-49
BE609806 weakly similar to PIR|T10521|T105 beta-glucosidase (EC ... 118 5e-27
TC213935 similar to UP|Q9SD73 (Q9SD73) Beta-D-glucan exohydrolas... 107 1e-23
CA935139 similar to PIR|T45640|T456 beta-D-glucan exohydrolase-l... 94 1e-19
TC225267 similar to UP|Q7XJH8 (Q7XJH8) Auxin-induced beta-glucos... 89 3e-18
TC228735 similar to UP|Q76MS5 (Q76MS5) LEXYL1 protein, partial (... 82 6e-16
BE612316 79 4e-15
BE191095 weakly similar to PIR|T45641|T456 beta-D-glucan exohydr... 73 2e-13
AW309079 similar to PIR|T51281|T512 beta-D-glucan exohydrolase (... 61 8e-10
BU549764 51 8e-07
BE805639 47 2e-05
TC232682 similar to UP|Q7XAS3 (Q7XAS3) Beta-D-glucosidase, parti... 47 2e-05
TC225271 similar to UP|Q6RXY3 (Q6RXY3) Beta xylosidase (Fragment... 44 1e-04
>TC206214 beta-D-glucan exohydrolase [Glycine max]
Length = 1647
Score = 606 bits (1563), Expect = e-174
Identities = 288/404 (71%), Positives = 341/404 (84%)
Frame = +1
Query: 2 KVIACAKHYVGDGGTINGIDGNNTVIDRDGLMRVHMPGYFSSISKGVATIMVSYSSWNGI 61
KV ACAKHY+GDGGT GI+ NNT+I +GL+ +HMP Y+ SI KGV+T+M+SYSSWNG+
Sbjct: 277 KVAACAKHYLGDGGTNKGINENNTLISYNGLLSIHMPAYYDSIIKGVSTVMISYSSWNGM 456
Query: 62 KMHTNHDMITGFLKNTLHFKGFVISDFEGIDRITSPPHANFTYSVEAGVSAGIDMFMIPK 121
KMH N +ITG+LKN LHFKGFVISD++GIDRITSPPHAN++YSV+AGVSAGIDM M+P
Sbjct: 457 KMHANKKLITGYLKNKLHFKGFVISDWQGIDRITSPPHANYSYSVQAGVSAGIDMIMVPF 636
Query: 122 FFTEFINDLTILVKNKLIPMSRIDDAVKRILWVKFMMGIFENPFADYSLVGYLGIQKHRK 181
+TEFI++LT VKN +IP+SRIDDAV RIL VKF+MG+FENP+AD SL LG ++HR+
Sbjct: 637 NYTEFIDELTRQVKNNIIPISRIDDAVARILRVKFVMGLFENPYADPSLANQLGSKEHRE 816
Query: 182 LAREAVRKSLVLLKNGISAEKPILPLTKKVPKILVAGTHADNLGYQCGGWTIEWQGVSGN 241
+AREAVRKSLVLLKNG S +KP+LPL KK KILVAG+HA+NLGYQCGGWTI WQG+ GN
Sbjct: 817 IAREAVRKSLVLLKNGKSYKKPLLPLPKKSTKILVAGSHANNLGYQCGGWTITWQGLGGN 996
Query: 242 NHLEGTTILTAIKNTVDPETTVIYKENPDTEFVESNGFSYAIVVVGEHPYAEMEGDSMNL 301
+ GTTIL A+K TVDP T V++ ENPD FV+S F YAIVVVGEH YAE GDS+NL
Sbjct: 997 DLTSGTTILDAVKQTVDPATEVVFNENPDKNFVKSYKFDYAIVVVGEHTYAETFGDSLNL 1176
Query: 302 TIPSPGPETITNVCEAITCVVIIISGRPLVIEPYLALIDALVAGWLPGSEGQGVADVLYG 361
T+ PGP TITNVC AI CVV++++GRP+VI+PYL IDALVA WLPG+EGQGVADVLYG
Sbjct: 1177TMADPGPSTITNVCGAIRCVVVLVTGRPVVIKPYLPKIDALVAAWLPGTEGQGVADVLYG 1356
Query: 362 DYGFTGKLPRTWFKSVDQLPMNVGDPHYDPLFPFGFGLSTQPTK 405
DY FTGKL RTWFK+VDQLPMNVGD HYDPLFPFG+GL+T TK
Sbjct: 1357DYEFTGKLARTWFKTVDQLPMNVGDKHYDPLFPFGYGLTTNITK 1488
>TC228369 similar to UP|Q7XAS3 (Q7XAS3) Beta-D-glucosidase, partial (49%)
Length = 1356
Score = 374 bits (960), Expect = e-104
Identities = 178/246 (72%), Positives = 200/246 (80%)
Frame = +2
Query: 160 IFENPFADYSLVGYLGIQKHRKLAREAVRKSLVLLKNGISAEKPILPLTKKVPKILVAGT 219
+FENP AD SLV LG Q+HR LAREAVRKSLVLLKNG + +LPL KKVPKILVAG+
Sbjct: 206 LFENPLADTSLVNELGSQEHRDLAREAVRKSLVLLKNGKNESASLLPLPKKVPKILVAGS 385
Query: 220 HADNLGYQCGGWTIEWQGVSGNNHLEGTTILTAIKNTVDPETTVIYKENPDTEFVESNGF 279
HADNLGYQCGGWTI+WQG SGN+ GTTIL AIK+ VD T V++++NPD EFV SN F
Sbjct: 386 HADNLGYQCGGWTIKWQGFSGNSDTRGTTILNAIKSAVDTSTEVVFRDNPDNEFVRSNNF 565
Query: 280 SYAIVVVGEHPYAEMEGDSMNLTIPSPGPETITNVCEAITCVVIIISGRPLVIEPYLALI 339
YAIVVVGE PYAE GDS L + GP I NVC + CVV+IISGRP+VIEPY++ I
Sbjct: 566 EYAIVVVGEPPYAETAGDSTTLAMMESGPNVINNVCGTVKCVVVIISGRPIVIEPYVSSI 745
Query: 340 DALVAGWLPGSEGQGVADVLYGDYGFTGKLPRTWFKSVDQLPMNVGDPHYDPLFPFGFGL 399
DALVA WLPG+EGQGV DVL+GDYGFTGKL RTWFKSVDQLPMNVGDPHYDPLFPFGFGL
Sbjct: 746 DALVAAWLPGTEGQGVTDVLFGDYGFTGKLARTWFKSVDQLPMNVGDPHYDPLFPFGFGL 925
Query: 400 STQPTK 405
+T+ K
Sbjct: 926 TTESVK 943
Score = 92.8 bits (229), Expect = 2e-19
Identities = 45/79 (56%), Positives = 59/79 (73%)
Frame = +1
Query: 92 DRITSPPHANFTYSVEAGVSAGIDMFMIPKFFTEFINDLTILVKNKLIPMSRIDDAVKRI 151
DR+TSPP +N+TYSV+A + AG+DM M+P + +FI DLT+LVK+ +IPM RIDDAV+RI
Sbjct: 1 DRLTSPPSSNYTYSVQASIEAGVDMVMVPFEYDKFIQDLTLLVKSNIIPMERIDDAVERI 180
Query: 152 LWVKFMMGIFENPFADYSL 170
L VKF M F + Y L
Sbjct: 181 LLVKFTMLSF*ESSS*YQL 237
>TC224183 similar to UP|Q7XAS3 (Q7XAS3) Beta-D-glucosidase, partial (29%)
Length = 542
Score = 324 bits (830), Expect = 5e-89
Identities = 153/179 (85%), Positives = 167/179 (92%)
Frame = +2
Query: 2 KVIACAKHYVGDGGTINGIDGNNTVIDRDGLMRVHMPGYFSSISKGVATIMVSYSSWNGI 61
KVI CAKHYVGDGGT NGID +NTVIDRDGLM++HMPGYFSSISKGVATIM SYSSWNG+
Sbjct: 2 KVIGCAKHYVGDGGTTNGIDEHNTVIDRDGLMKIHMPGYFSSISKGVATIMASYSSWNGV 181
Query: 62 KMHTNHDMITGFLKNTLHFKGFVISDFEGIDRITSPPHANFTYSVEAGVSAGIDMFMIPK 121
KMH +HD+ITG+LKNTLHFKGFVISDFEGIDRITSPP AN TYS+EAGVSAGIDMFM+PK
Sbjct: 182 KMHAHHDLITGYLKNTLHFKGFVISDFEGIDRITSPPRANITYSIEAGVSAGIDMFMVPK 361
Query: 122 FFTEFINDLTILVKNKLIPMSRIDDAVKRILWVKFMMGIFENPFADYSLVGYLGIQKHR 180
+TEFI+ LT+LVKNK IPMSRIDDAV+RILWVKFMMGIFENPFADYSL YLGIQ+HR
Sbjct: 362 HYTEFIDVLTMLVKNKHIPMSRIDDAVRRILWVKFMMGIFENPFADYSLAKYLGIQEHR 538
>TC229253 similar to UP|Q7XAS3 (Q7XAS3) Beta-D-glucosidase, partial (26%)
Length = 601
Score = 311 bits (798), Expect = 3e-85
Identities = 147/170 (86%), Positives = 158/170 (92%)
Frame = +3
Query: 240 GNNHLEGTTILTAIKNTVDPETTVIYKENPDTEFVESNGFSYAIVVVGEHPYAEMEGDSM 299
GNN L+GTTIL A+KNTVDPETTV+YKENPD EFV+SN FSYAIVVVGEHPYAEM GDSM
Sbjct: 3 GNNLLKGTTILAAVKNTVDPETTVVYKENPDVEFVKSNEFSYAIVVVGEHPYAEMYGDSM 182
Query: 300 NLTIPSPGPETITNVCEAITCVVIIISGRPLVIEPYLALIDALVAGWLPGSEGQGVADVL 359
NLTIP PGP+ ITNVC AI CVVIIISGRP+VIEPY+ LIDALVA WLPGSEGQGVADVL
Sbjct: 183 NLTIPEPGPKIITNVCGAIKCVVIIISGRPVVIEPYVGLIDALVAAWLPGSEGQGVADVL 362
Query: 360 YGDYGFTGKLPRTWFKSVDQLPMNVGDPHYDPLFPFGFGLSTQPTKAIYS 409
YG YGFTGKLPRTWFK+VDQLPMNVGDPHYDPLFPFGFGLST+P+KA+YS
Sbjct: 363 YGGYGFTGKLPRTWFKTVDQLPMNVGDPHYDPLFPFGFGLSTKPSKALYS 512
>TC206215 similar to UP|Q7XAS3 (Q7XAS3) Beta-D-glucosidase, partial (27%)
Length = 673
Score = 277 bits (708), Expect = 7e-75
Identities = 126/170 (74%), Positives = 147/170 (86%)
Frame = +1
Query: 236 QGVSGNNHLEGTTILTAIKNTVDPETTVIYKENPDTEFVESNGFSYAIVVVGEHPYAEME 295
QG+ GNN GTTIL A+K T+DP T V+Y ENPD+ FV+SN FSYAIV+VGEHPYA+
Sbjct: 1 QGLGGNNLTVGTTILEAVKQTIDPATKVVYNENPDSNFVKSNNFSYAIVIVGEHPYAKTF 180
Query: 296 GDSMNLTIPSPGPETITNVCEAITCVVIIISGRPLVIEPYLALIDALVAGWLPGSEGQGV 355
GDS+NLTIP PGP TITNVC +I CVV++I+GRP+VI+PYL+ IDALVA WLPG+EGQGV
Sbjct: 181 GDSLNLTIPEPGPSTITNVCGSIQCVVVLITGRPVVIQPYLSKIDALVAAWLPGTEGQGV 360
Query: 356 ADVLYGDYGFTGKLPRTWFKSVDQLPMNVGDPHYDPLFPFGFGLSTQPTK 405
D+L+GDYGFTGKL RTWFK+VDQLPMNVGD +YDPLFPFGFGLST PTK
Sbjct: 361 TDLLFGDYGFTGKLARTWFKTVDQLPMNVGDKYYDPLFPFGFGLSTNPTK 510
>BF066868 weakly similar to PIR|T51283|T51 glucan 1 3-beta-glucosidase (EC
3.2.1.58) [imported] - common tobacco, partial (30%)
Length = 595
Score = 258 bits (660), Expect = 3e-69
Identities = 125/193 (64%), Positives = 146/193 (74%)
Frame = +2
Query: 209 KKVPKILVAGTHADNLGYQCGGWTIEWQGVSGNNHLEGTTILTAIKNTVDPETTVIYKEN 268
KK KILVAG+HADNLGYQCGGWTI QG GNN TIL A+K +DP T V+Y EN
Sbjct: 14 KKAAKILVAGSHADNLGYQCGGWTITCQGGGGNNLTVCATILDAVKQAIDPATKVVYNEN 193
Query: 269 PDTEFVESNGFSYAIVVVGEHPYAEMEGDSMNLTIPSPGPETITNVCEAITCVVIIISGR 328
PD+ FV+SN FSYAIV VGEHPYAE GDS+NLTI PGP TITNVC +I CVV++I+ R
Sbjct: 194 PDSNFVKSNNFSYAIVTVGEHPYAETFGDSLNLTISEPGPSTITNVCGSIQCVVVLITCR 373
Query: 329 PLVIEPYLALIDALVAGWLPGSEGQGVADVLYGDYGFTGKLPRTWFKSVDQLPMNVGDPH 388
P+VI+PYL+ IDALVA WL +E Q V D+LYGDYG T TW+K+VDQL MNV D +
Sbjct: 374 PVVIQPYLSKIDALVAAWLTLTECQSVTDLLYGDYGCTV*YAITWYKTVDQLLMNVRDEY 553
Query: 389 YDPLFPFGFGLST 401
YDP FG+G+ST
Sbjct: 554 YDPCITFGYGIST 592
>AI973674 weakly similar to PIR|T10521|T10 beta-glucosidase (EC 3.2.1.21) -
common nasturtium, partial (22%)
Length = 457
Score = 198 bits (504), Expect = 3e-51
Identities = 98/150 (65%), Positives = 111/150 (73%)
Frame = +2
Query: 252 AIKNTVDPETTVIYKENPDTEFVESNGFSYAIVVVGEHPYAEMEGDSMNLTIPSPGPETI 311
A K DP T V++ ENPD FV+ F YAI V GEH YAE GDS+NLT+ PGP TI
Sbjct: 5 AEKQNDDPATEVLFNENPDINFVK*YKFDYAIDVEGEHTYAETFGDSLNLTMADPGPSTI 184
Query: 312 TNVCEAITCVVIIISGRPLVIEPYLALIDALVAGWLPGSEGQGVADVLYGDYGFTGKLPR 371
TNVC AI CV +++ RP VI+PYL IDALVA WLPG+EGQGVADVLYGDY FT KL R
Sbjct: 185 TNVCGAIRCVYVLVCCRPFVIKPYLPKIDALVAAWLPGTEGQGVADVLYGDYEFTRKLAR 364
Query: 372 TWFKSVDQLPMNVGDPHYDPLFPFGFGLST 401
T F +VDQLPMNV D HYDPLF FG+ L+T
Sbjct: 365 TCFMTVDQLPMNVEDKHYDPLFAFGYVLTT 454
>TC213093 similar to UP|Q7XAS3 (Q7XAS3) Beta-D-glucosidase, partial (21%)
Length = 406
Score = 191 bits (486), Expect = 4e-49
Identities = 96/134 (71%), Positives = 114/134 (84%)
Frame = +3
Query: 85 ISDFEGIDRITSPPHANFTYSVEAGVSAGIDMFMIPKFFTEFINDLTILVKNKLIPMSRI 144
ISD+ GID+ITSPPH+N++YS++ GV AGIDM M+P FTEFI+ LT VKN +IP+SRI
Sbjct: 3 ISDWLGIDKITSPPHSNYSYSIQVGVGAGIDMIMVPFNFTEFIDVLTYQVKNNIIPVSRI 182
Query: 145 DDAVKRILWVKFMMGIFENPFADYSLVGYLGIQKHRKLAREAVRKSLVLLKNGISAEKPI 204
DDAVKRIL VKF+MG+FENP AD SLV LG ++HR++AREAVRKSLVLLKNG SAEKP+
Sbjct: 183 DDAVKRILRVKFVMGLFENPLADLSLVNQLGSEEHRQIAREAVRKSLVLLKNGKSAEKPL 362
Query: 205 LPLTKKVPKILVAG 218
LPL KK KILVAG
Sbjct: 363 LPLPKKAAKILVAG 404
>BE609806 weakly similar to PIR|T10521|T105 beta-glucosidase (EC 3.2.1.21) -
common nasturtium, partial (12%)
Length = 238
Score = 118 bits (295), Expect = 5e-27
Identities = 59/79 (74%), Positives = 64/79 (80%)
Frame = +2
Query: 250 LTAIKNTVDPETTVIYKENPDTEFVESNGFSYAIVVVGEHPYAEMEGDSMNLTIPSPGPE 309
LTA+KNTVDPETTV+YKENPD FV+SNGFSYAI +VGEHPYAEM GDSM LT PGP
Sbjct: 2 LTAVKNTVDPETTVVYKENPDV*FVKSNGFSYAIDIVGEHPYAEMYGDSMILTFRKPGPR 181
Query: 310 TITNVCEAITCVVIIISGR 328
I V A+ CVVIIISGR
Sbjct: 182 IIAVVWGAL*CVVIIISGR 238
>TC213935 similar to UP|Q9SD73 (Q9SD73) Beta-D-glucan exohydrolase-like
protein (At3g47000), partial (16%)
Length = 903
Score = 107 bits (267), Expect = 1e-23
Identities = 55/106 (51%), Positives = 73/106 (67%), Gaps = 1/106 (0%)
Frame = +3
Query: 281 YAIVVVGEHPYAEMEGDSMNLTIPSPGPETITNVCEAITCVVIIISGRPLVIEP-YLALI 339
+AIV VGE PYAE GD+ LTIP G + I+ V + I +VI+ISGR LV+EP L I
Sbjct: 3 FAIVAVGEAPYAETWGDNSELTIPVNGADIISLVADQIPTLVILISGRRLVLEPRLLEKI 182
Query: 340 DALVAGWLPGSEGQGVADVLYGDYGFTGKLPRTWFKSVDQLPMNVG 385
DALVA WLPG+EG+G+ DV++ + F KLP TWF+ V++ +G
Sbjct: 183 DALVAAWLPGNEGEGIIDVIFCSHDFKDKLPVTWFRRVERQISQIG 320
>CA935139 similar to PIR|T45640|T456 beta-D-glucan exohydrolase-like protein
- Arabidopsis thaliana, partial (12%)
Length = 446
Score = 94.0 bits (232), Expect = 1e-19
Identities = 47/78 (60%), Positives = 58/78 (74%), Gaps = 2/78 (2%)
Frame = +2
Query: 325 ISGRPLVIEPYLA-LIDALVAGWLPGSEGQGVADVLYGDYGFTGKLPRTWFKSVDQLPMN 383
ISGRPLV+EP L IDALVA WLPGSEG+G+ DV++G +GF GKLP TWF+ V+QL
Sbjct: 2 ISGRPLVLEPLLLDKIDALVAVWLPGSEGEGITDVIFGSHGFKGKLPVTWFRRVEQLDQP 181
Query: 384 VGDPH-YDPLFPFGFGLS 400
+ +PLFP GFGL+
Sbjct: 182 ADAVNSCEPLFPLGFGLA 235
>TC225267 similar to UP|Q7XJH8 (Q7XJH8) Auxin-induced beta-glucosidase,
partial (69%)
Length = 2006
Score = 89.4 bits (220), Expect = 3e-18
Identities = 114/446 (25%), Positives = 195/446 (43%), Gaps = 47/446 (10%)
Frame = +3
Query: 2 KVIACAKHYVG-DGGTINGID--GNNTVIDRDGLMRVHMPGYFSSISKG-VATIMVSYSS 57
KV AC KHY D NG+D N + + L + + + + +G VA++M SY+
Sbjct: 30 KVAACCKHYTAYDLDNWNGVDRFHFNAKVSKQDLEDTYDVPFKACVLEGQVASVMCSYNQ 209
Query: 58 WNGIKMHTNHDMITGFLKNTLHFKGFVISDFEGIDRITSPPHANFTYSVEA--GVSAGID 115
NG + D++ ++ G+++SD + + H T A + AG+D
Sbjct: 210 VNGKPTCADPDLLRNTIRGQWRLNGYIVSDCDSVGVFFDNQHYTKTPEEAAAEAIKAGLD 389
Query: 116 MFMIPKFFTEFINDLTILVKNKLIPMSRIDDAVKRILWVKFMMGIF--ENPFADYSLVGY 173
+ P F D I + LI + ++ A+ ++ V+ +G+F E Y +G
Sbjct: 390 LDCGP--FLAIHTDSAI--RKGLISENDLNLALANLISVQMRLGMFDGEPSTQPYGNLGP 557
Query: 174 LGI--QKHRKLAREAVRKSLVLLKNGISAEKPILPLT-KKVPKILVAGTHAD----NLGY 226
+ H++LA EA R+S+VLL+N ++ LPL+ ++ I V G +AD +G
Sbjct: 558 RDVCTSAHQQLALEAARESIVLLQNKGNS----LPLSPSRLRTIGVVGPNADATVTMIGN 725
Query: 227 QCG---GWTIEWQGVS---GNNHLEGTTILTAIKNTVDPETTVIYKENPDTEFVESNGFS 280
G G+T QG++ H G + N + I ++
Sbjct: 726 YAGVACGYTTPLQGIARYVKTAHQVGCKGVACRGNELFGAAETIARQ-----------AD 872
Query: 281 YAIVVVGEHPYAEME-GDSMNLTIPSPGPETITNVCEAI--TCVVIIISGRPLVI----- 332
++V+G E E D + L +P E +T V A +++I+SG P+ I
Sbjct: 873 AIVLVMGLDQTVEAETRDRVGLLLPGLQQELVTRVARAAKGPVILLIMSGGPVDISFAKN 1052
Query: 333 EPYLALIDALVAGWLPGSEGQGVADVLYGDYGFTGKLPRTWFKS--VDQLPMNVGDPHYD 390
+P ++ I L G+ + G +ADV++G G+LP TW+ + ++PM D +
Sbjct: 1053DPKISAI--LWVGYPGQAGGTAIADVIFGTTNPGGRLPMTWYPQGYLAKVPMTNMDMRPN 1226
Query: 391 P----------------LFPFGFGLS 400
P +FPFG GLS
Sbjct: 1227PTTGYPGRTYRFYKGPVVFPFGHGLS 1304
>TC228735 similar to UP|Q76MS5 (Q76MS5) LEXYL1 protein, partial (65%)
Length = 1599
Score = 81.6 bits (200), Expect = 6e-16
Identities = 97/389 (24%), Positives = 173/389 (43%), Gaps = 36/389 (9%)
Frame = +1
Query: 48 VATIMVSYSSWNGIKMHTNHDMITGFLKNTLHFKGFVISDFEGIDRITSPPHANFTYSVE 107
VA++M SY+ NG + D++ G ++ G+++SD + ++ + H T
Sbjct: 1 VASVMCSYNKVNGKPTCADPDLLKGVVRGEWKLNGYIVSDCDSVEVLYKDQHYTKTPEEA 180
Query: 108 AGVS--AGIDMFMIPKFFTEFINDLTILVKNKLIPMSRIDDAVKRILWVKFMMGIFENP- 164
A +S AG+D+ +F ++ VK LI + I++AV +G F+
Sbjct: 181 AAISILAGLDL-NCGRFLGQYTEGA---VKQGLIDEASINNAVTNNFATLMRLGFFDGDP 348
Query: 165 -FADYSLVGYLGI--QKHRKLAREAVRKSLVLLKNGISAEKPILPLTKKVPKIL-VAGTH 220
Y +G + Q++++LAREA R+ +VLLKN ++ LPL K K L V G +
Sbjct: 349 RKQPYGNLGPKDVCTQENQELAREAARQGIVLLKNSPAS----LPLNAKAIKSLAVIGPN 516
Query: 221 ADNLGYQCGGWTIEWQGVSGN--NHLEGTTILTAIKNTVDPETTVIYKENP---DTEFVE 275
A+ G ++G+ + L+G T + NP D + +
Sbjct: 517 ANATRVMIG----NYEGIPCKYISPLQGLTAFAPTSYAAG--CLDVRCPNPVLDDAKKIA 678
Query: 276 SNGFSYAIVVVGEHPYAEMEG-DSMNLTIPSPGPETITNVCEAI--TCVVIIISGRPLVI 332
++ ++VVG E E D +N+ +P ++ V A +++I+SG + +
Sbjct: 679 ASA-DATVMVVGASLAIEAESLDRVNILLPGQQQLLVSEVANASKGPVILVIMSGGGMDV 855
Query: 333 ---EPYLALIDALVAGWLPGSEGQGVADVLYGDYGFTGKLPRTWFKS--VDQLPMNVGDP 387
+ + L G+ + G +ADV++G + +G+LP TW+ VD++PM +
Sbjct: 856 SFAKNNNKITSILWVGYPGEAGGAAIADVIFGFHNPSGRLPMTWYPQSYVDKVPMTNMNM 1035
Query: 388 HYDP----------------LFPFGFGLS 400
DP +F FG GLS
Sbjct: 1036RPDPATGYPGRTYRFYKGETVFAFGDGLS 1122
>BE612316
Length = 181
Score = 79.0 bits (193), Expect = 4e-15
Identities = 35/51 (68%), Positives = 41/51 (79%)
Frame = +2
Query: 36 HMPGYFSSISKGVATIMVSYSSWNGIKMHTNHDMITGFLKNTLHFKGFVIS 86
HMP Y SI KGV+T+MVSYSSWNG KMH NH ++T +LKN L+F GFVIS
Sbjct: 2 HMPAYHDSIVKGVSTVMVSYSSWNGQKMHANHFLVTDYLKNKLNFMGFVIS 154
>BE191095 weakly similar to PIR|T45641|T456 beta-D-glucan exohydrolase-like
protein - Arabidopsis thaliana, partial (15%)
Length = 328
Score = 73.2 bits (178), Expect = 2e-13
Identities = 43/101 (42%), Positives = 58/101 (56%), Gaps = 1/101 (0%)
Frame = +2
Query: 286 VGEHPYAEMEGDSMNLTIPSPGPETITNVCEAITCVVIIISGRPLVIEPY-LALIDALVA 344
VGE YAE G LTIP+ G + I I +VI+ISGR ++ P L IDAL A
Sbjct: 11 VGEAAYAEDLG*QYQLTIPANGADIINLYANQIPTMVILISGRR*LL*PQILENIDALDA 190
Query: 345 GWLPGSEGQGVADVLYGDYGFTGKLPRTWFKSVDQLPMNVG 385
W+P +EG G+ V++ + F KLP+TWFK V+ +G
Sbjct: 191 AWVPRNEG*GIIYVIFCKHDFKDKLPKTWFKRVEPTISQIG 313
>AW309079 similar to PIR|T51281|T512 beta-D-glucan exohydrolase (EC
3.2.1.-) isoenzyme ExoI [imported] - barley, partial
(5%)
Length = 414
Score = 61.2 bits (147), Expect = 8e-10
Identities = 26/39 (66%), Positives = 33/39 (83%), Gaps = 2/39 (5%)
Frame = +1
Query: 49 ATIMVSYSSWNGIKMHTNHDMITGFLKNTLHFK--GFVI 85
+T+MVSYSSWNG++MH N D++TGFLKNTL FK GF +
Sbjct: 1 STVMVSYSSWNGVRMHANRDLVTGFLKNTLKFKVIGFFV 117
>BU549764
Length = 479
Score = 51.2 bits (121), Expect = 8e-07
Identities = 22/36 (61%), Positives = 27/36 (74%)
Frame = -2
Query: 2 KVIACAKHYVGDGGTINGIDGNNTVIDRDGLMRVHM 37
KV ACAKHYVGD GT GI+ NN V +GL+R+H+
Sbjct: 160 KVAACAKHYVGDDGTTKGINENNIVASYNGLLRIHI 53
>BE805639
Length = 419
Score = 47.0 bits (110), Expect = 2e-05
Identities = 34/105 (32%), Positives = 47/105 (44%), Gaps = 4/105 (3%)
Frame = +1
Query: 281 YAIVVVGEHPYAEMEG-DSMNLTIPSPGPET--ITNVCEAITCVVIIISGRPLVIEPYLA 337
YA+V G E EG D ++ +P G + I V A T V+I S + P+L
Sbjct: 40 YALVFTGHTTSWETEGQDQLSFNLPKDGSQDRLIAGVAAANTQTVVINSTGVAIAMPWLE 219
Query: 338 LIDALVAGWLPGSE-GQGVADVLYGDYGFTGKLPRTWFKSVDQLP 381
I L+ W PG E G + DVL G G L ++ K + P
Sbjct: 220 EISGLLQTWFPGQEAGNSIVDVLTGKVNAEGALTCSFPKELTSAP 354
>TC232682 similar to UP|Q7XAS3 (Q7XAS3) Beta-D-glucosidase, partial (5%)
Length = 526
Score = 46.6 bits (109), Expect = 2e-05
Identities = 21/33 (63%), Positives = 24/33 (72%)
Frame = +2
Query: 2 KVIACAKHYVGDGGTINGIDGNNTVIDRDGLMR 34
KV ACAKHYVGD GT GI+ NN V +GL+R
Sbjct: 386 KVAACAKHYVGDDGTTKGINENNIVASYNGLLR 484
>TC225271 similar to UP|Q6RXY3 (Q6RXY3) Beta xylosidase (Fragment), partial
(55%)
Length = 952
Score = 44.3 bits (103), Expect = 1e-04
Identities = 40/144 (27%), Positives = 66/144 (45%), Gaps = 26/144 (18%)
Frame = +3
Query: 283 IVVVGEHPYAEMEG-DSMNLTIPSPGPETITNVCEAIT--CVVIIISGRPLVIE-----P 334
++V+G E E D + L +P E +T V A +++I+SG P+ + P
Sbjct: 9 VLVMGLDQTIEAETRDRVGLLLPGLQQELVTRVARAAKGPVILVIMSGGPVDVSFAKNNP 188
Query: 335 YLALIDALVAGWLPGSEGQGVADVLYGDYGFTGKLPRTWFKS--VDQLPMNVGDPHYDP- 391
++ I L G+ + G +ADV++G G+LP TW+ + ++PM D +P
Sbjct: 189 KISAI--LWVGYPGQAGGTAIADVIFGATNPGGRLPMTWYPQGYLAKVPMTNMDMRPNPA 362
Query: 392 ---------------LFPFGFGLS 400
+FPFG GLS
Sbjct: 363 TGYPGRTYRFYKGPVVFPFGHGLS 434
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.320 0.140 0.427
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 18,337,800
Number of Sequences: 63676
Number of extensions: 251876
Number of successful extensions: 1055
Number of sequences better than 10.0: 51
Number of HSP's better than 10.0 without gapping: 1047
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1051
length of query: 409
length of database: 12,639,632
effective HSP length: 100
effective length of query: 309
effective length of database: 6,272,032
effective search space: 1938057888
effective search space used: 1938057888
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0371a.1


