

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0348.15
(242 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
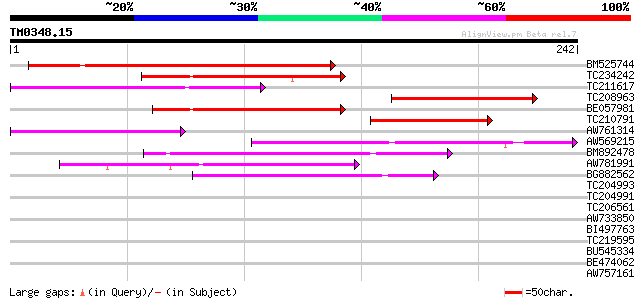
Score E
Sequences producing significant alignments: (bits) Value
BM525744 124 3e-29
TC234242 82 3e-16
TC211617 80 8e-16
TC208963 64 5e-11
BE057981 63 1e-10
TC210791 53 1e-07
AW761314 53 1e-07
AW569215 weakly similar to GP|4914326|gb| F14N23.12 {Arabidopsis... 52 2e-07
BM892478 51 5e-07
AW781991 48 5e-06
BG882562 46 1e-05
TC204993 similar to UP|Q8H146 (Q8H146) Putatative tyrosine amino... 30 0.75
TC204991 similar to UP|Q9FN30 (Q9FN30) Tyrosine aminotransferase... 30 0.98
TC206561 29 2.2
AW733850 28 2.9
BI497763 28 2.9
TC219595 similar to UP|Q948R8 (Q948R8) THH1, partial (75%) 28 2.9
BU545334 28 4.9
BE474062 similar to GP|9758410|dbj lysine decarboxylase-like pro... 27 8.3
AW757161 27 8.3
>BM525744
Length = 411
Score = 124 bits (312), Expect = 3e-29
Identities = 63/131 (48%), Positives = 88/131 (67%)
Frame = +3
Query: 9 VDSMNNNWVPPRHMLATLKENNPGNLSTITQVYNRIKKVKELDCGPLTEMQYLLKKLAEA 68
V M V PR++L TLK++N N +TI +YN + + GP TEMQ+LLK L
Sbjct: 9 VIDMTKKMVEPRNILLTLKDHN--NDTTIRHIYNARQAYRSSQKGPRTEMQHLLKLLEHD 182
Query: 69 NYVHFKRHEEDSGVIMELFWSHPNAIKLFNTFPHVVIMDCTYKTNKFQIPLLEMVGLTST 128
YV + R +DS I ++FW+HP+AIKL +F V+ +D TYK N++Q+PLLE+VG+TST
Sbjct: 183 QYVCWSRKVDDSDAIRDIFWAHPDAIKLLGSFHTVLFLDNTYKVNRYQLPLLEIVGVTST 362
Query: 129 GLTYSIAFCYM 139
LT+S+AF YM
Sbjct: 363 ELTFSVAFAYM 395
>TC234242
Length = 478
Score = 81.6 bits (200), Expect = 3e-16
Identities = 38/92 (41%), Positives = 62/92 (67%), Gaps = 5/92 (5%)
Frame = +3
Query: 57 EMQYLLKKLAEANYVHFKRHEEDSGVIMELFWSHPNAIKLFNTFPHVVIMDCTYKTNKFQ 116
EMQ+L+K L Y+H+ R + D V+ ++FW HP+++KL N V ++D TY+TN+++
Sbjct: 15 EMQHLMKFLERDQYIHWHRIK-DEDVVRDIFWCHPDSVKLVNACNLVFLIDSTYRTNRYR 191
Query: 117 IPL-----LEMVGLTSTGLTYSIAFCYMTRER 143
+PL L+ VG+T TG+T+S +F Y+ ER
Sbjct: 192 LPLLDFVVLDFVGMTPTGMTFSASFAYVEGER 287
>TC211617
Length = 534
Score = 80.1 bits (196), Expect = 8e-16
Identities = 43/109 (39%), Positives = 65/109 (59%)
Frame = -1
Query: 1 LTEEEKVQVDSMNNNWVPPRHMLATLKENNPGNLSTITQVYNRIKKVKELDCGPLTEMQY 60
LT++EK + M + V PR++ TLKE+N + +TI Q+YN G TEMQ+
Sbjct: 333 LTDDEKNIIADMTKSNVKPRNIQLTLKEHNTNSCTTIKQIYNARSAYHSSIRGDDTEMQH 154
Query: 61 LLKKLAEANYVHFKRHEEDSGVIMELFWSHPNAIKLFNTFPHVVIMDCT 109
L++ L Y+H+ R +D V+ +LFW HP+A+KL NT V ++D T
Sbjct: 153 LMRLLKRDQYIHWHR-LKDQDVVRDLFWCHPDAVKLCNTCHLVFLIDST 10
>TC208963
Length = 1163
Score = 64.3 bits (155), Expect = 5e-11
Identities = 31/62 (50%), Positives = 45/62 (72%)
Frame = +1
Query: 164 LPGVIVTDRELALLSAVRNIFPGATHLLCLFHINKNVEAKCKLWVDTTDFKALEMQKWNE 223
+P VIVT ++AL+SAV+ +FP +++LLC FHIN+NV+AKCK V + + L M W+
Sbjct: 19 MPQVIVTVGDIALMSAVQVVFPSSSNLLCRFHINQNVKAKCKSIVHLKEKQELMMDAWDV 198
Query: 224 VV 225
VV
Sbjct: 199 VV 204
>BE057981
Length = 268
Score = 62.8 bits (151), Expect = 1e-10
Identities = 29/82 (35%), Positives = 50/82 (60%)
Frame = +2
Query: 62 LKKLAEANYVHFKRHEEDSGVIMELFWSHPNAIKLFNTFPHVVIMDCTYKTNKFQIPLLE 121
+K Y+H+ R + D V+ +LFW H +A+KL N V +++ YKTN++++ LL+
Sbjct: 2 MKLFERDQYIHWHRLK-DEFVVRDLFWCHLDAVKLCNACHLVFLINNAYKTNRYRLLLLD 178
Query: 122 MVGLTSTGLTYSIAFCYMTRER 143
G+T +T+S+ F Y+ ER
Sbjct: 179 FAGVTPARMTFSVGFAYLEGER 244
>TC210791
Length = 456
Score = 53.1 bits (126), Expect = 1e-07
Identities = 23/52 (44%), Positives = 39/52 (74%)
Frame = +1
Query: 155 KSLLADPARLPGVIVTDRELALLSAVRNIFPGATHLLCLFHINKNVEAKCKL 206
+ L+ +P VI+T R++AL+ AV+ +FP +++LLC FHI+KNV+AK ++
Sbjct: 1 RGLIFKDNEMPPVIITVRDIALMDAVQVVFPSSSNLLCRFHISKNVKAKSQV 156
>AW761314
Length = 331
Score = 52.8 bits (125), Expect = 1e-07
Identities = 26/75 (34%), Positives = 44/75 (58%)
Frame = +1
Query: 1 LTEEEKVQVDSMNNNWVPPRHMLATLKENNPGNLSTITQVYNRIKKVKELDCGPLTEMQY 60
LT++E + + +M + V P+++L TLKE+N + + I Q+YN G T+MQ
Sbjct: 106 LTKDENIIITNMTKSMVKPKNILLTLKEHNVNSYTAIKQIYNARSAYCSSIRGSDTKMQQ 285
Query: 61 LLKKLAEANYVHFKR 75
L+K L + Y+H+ R
Sbjct: 286 LMKLLEQDQYIHWHR 330
>AW569215 weakly similar to GP|4914326|gb| F14N23.12 {Arabidopsis thaliana},
partial (19%)
Length = 425
Score = 52.0 bits (123), Expect = 2e-07
Identities = 35/141 (24%), Positives = 61/141 (42%), Gaps = 2/141 (1%)
Frame = +1
Query: 104 VIMDCTYKTNKFQIPLLEMVGLTSTGLTYSIAFCYMTRERTPDYVWALECMKSLLADPAR 163
V+ D TY+ + + L +G+ + G+T + + E + WAL+ + A
Sbjct: 10 VVFDTTYRVEAYDMLLGIWLGVDNNGMTCFFSCALLRDENIQSFSWALKAFLGFMKGKA- 186
Query: 164 LPGVIVTDRELALLSAVRNIFPGATHLLCLFHINKNVEAKCKLWVDT--TDFKALEMQKW 221
P I+TD + L A+ P H C++HI L + + ++KA ++
Sbjct: 187 -PQTILTDHNMWLKEAIAVELPQTKHAFCIWHILSKFSDWFSLLLGSQYDEWKA----EF 351
Query: 222 NEVVYAETTTQFEEEWRDMCD 242
+ + E FEE WR M D
Sbjct: 352 HRLYNLEQVEDFEEGWRQMVD 414
>BM892478
Length = 429
Score = 50.8 bits (120), Expect = 5e-07
Identities = 30/132 (22%), Positives = 59/132 (43%)
Frame = +3
Query: 58 MQYLLKKLAEANYVHFKRHEEDSGVIMELFWSHPNAIKLFNTFPHVVIMDCTYKTNKFQI 117
++Y + AE + F E D+G M +FW+ + + F V+++D +Y+ + +
Sbjct: 33 LEYFQSRQAE-DTGFFYAMEVDNGNCMNIFWADGRSRYSCSHFGDVLVLDTSYRKTVYLV 209
Query: 118 PLLEMVGLTSTGLTYSIAFCYMTRERTPDYVWALECMKSLLADPARLPGVIVTDRELALL 177
P VG+ + + E + W + L A RLP ++ D+++A+
Sbjct: 210 PFATFVGVNHHKQPVLLGCALIADESEESFTWLFQTW--LRAMSGRLPLTVIADQDIAIQ 383
Query: 178 SAVRNIFPGATH 189
A+ +FP H
Sbjct: 384 RAIAKVFPVTHH 419
>AW781991
Length = 419
Score = 47.8 bits (112), Expect = 5e-06
Identities = 33/133 (24%), Positives = 59/133 (43%), Gaps = 5/133 (3%)
Frame = +3
Query: 22 MLATLKENNPGNLSTITQV----YNRIKKVKELDCGPLTEMQYLLKKLAE-ANYVHFKRH 76
M A +KE + T+V Y R +++ L+ + YL + AE N+ + +
Sbjct: 9 MSALIKEYGGISKVGFTEVDCRNYMRNNRLRSLEGDIQLVLDYLRQMHAENPNFFYAVQG 188
Query: 77 EEDSGVIMELFWSHPNAIKLFNTFPHVVIMDCTYKTNKFQIPLLEMVGLTSTGLTYSIAF 136
+ED I +FW+ P A + F V D TY++N++++P G+ G
Sbjct: 189 DEDQS-ITNVFWADPKARMNYTFFGDTVTFDTTYRSNRYRLPFAFFTGVNHHGQPVLFGC 365
Query: 137 CYMTRERTPDYVW 149
++ E +VW
Sbjct: 366 AFLINESEASFVW 404
>BG882562
Length = 416
Score = 46.2 bits (108), Expect = 1e-05
Identities = 26/105 (24%), Positives = 47/105 (44%)
Frame = +3
Query: 79 DSGVIMELFWSHPNAIKLFNTFPHVVIMDCTYKTNKFQIPLLEMVGLTSTGLTYSIAFCY 138
D G + +FW + ++ F VV D T +N ++IPL+ VG+ G + +
Sbjct: 108 DDGQLRNVFWIDSRSRAAYSYFGDVVAFDSTCLSNNYEIPLVAFVGVNHHGKSVLLGCGL 287
Query: 139 MTRERTPDYVWALECMKSLLADPARLPGVIVTDRELALLSAVRNI 183
+ E Y+W + + R P I+T+ A+ SA+ +
Sbjct: 288 LADETFETYIWLFRAWLTCMT--GRPPQTIITNXCKAMQSAIAEV 416
>TC204993 similar to UP|Q8H146 (Q8H146) Putatative tyrosine aminotransferase,
partial (43%)
Length = 801
Score = 30.4 bits (67), Expect = 0.75
Identities = 27/129 (20%), Positives = 52/129 (39%), Gaps = 1/129 (0%)
Frame = +3
Query: 11 SMNNNWVPPRHMLATLKENNPGNLSTITQVYNRIKKVKELDCGPLTEMQYLLKKLAE-AN 69
S + W+ P L N+P +V R KK +L GP T +Q + ++ E
Sbjct: 84 SFSKRWIVPGWRLGWFVTNDPSGTFRNPKVDERFKKYFDLLGGPATFIQAAVPQIIEHTE 263
Query: 70 YVHFKRHEEDSGVIMELFWSHPNAIKLFNTFPHVVIMDCTYKTNKFQIPLLEMVGLTSTG 129
V FK+ ++ + ++ K P+++ C YK ++++
Sbjct: 264 KVFFKKTIDNLRHVADI------CCKELKDIPYII---CPYKPEGSMAMMVKLNLSLLED 416
Query: 130 LTYSIAFCY 138
++ I FC+
Sbjct: 417 ISDDIDFCF 443
>TC204991 similar to UP|Q9FN30 (Q9FN30) Tyrosine aminotransferase, partial
(89%)
Length = 1743
Score = 30.0 bits (66), Expect = 0.98
Identities = 18/66 (27%), Positives = 31/66 (46%), Gaps = 1/66 (1%)
Frame = +2
Query: 11 SMNNNWVPPRHMLATLKENNPGNLSTITQVYNRIKKVKELDCGPLTEMQYLLKK-LAEAN 69
S++ W+ P L N+P +V RIKK +L GP T +Q + + +A
Sbjct: 1022 SLSKRWIVPGWRLGWFVTNDPSGTFREPKVVERIKKYFDLLGGPATFLQAAVPQIIANTE 1201
Query: 70 YVHFKR 75
+ F++
Sbjct: 1202 EIFFEK 1219
>TC206561
Length = 1637
Score = 28.9 bits (63), Expect = 2.2
Identities = 12/39 (30%), Positives = 23/39 (58%)
Frame = -2
Query: 182 NIFPGATHLLCLFHINKNVEAKCKLWVDTTDFKALEMQK 220
N+F T++ F++N+ +EA K++ TDF + +K
Sbjct: 1561 NVFRLYTNIKNFFYMNEEIEAVHKIYYSETDFSCTDKRK 1445
>AW733850
Length = 424
Score = 28.5 bits (62), Expect = 2.9
Identities = 15/44 (34%), Positives = 25/44 (56%)
Frame = +3
Query: 162 ARLPGVIVTDRELALLSAVRNIFPGATHLLCLFHINKNVEAKCK 205
ARLP IV RE ++L +RN+ HL + ++N +++ K
Sbjct: 48 ARLPRHIVRPREPSILPQLRNLRHHRLHLRSICNLNSELKSHSK 179
>BI497763
Length = 421
Score = 28.5 bits (62), Expect = 2.9
Identities = 12/24 (50%), Positives = 16/24 (66%)
Frame = +3
Query: 19 PRHMLATLKENNPGNLSTITQVYN 42
PR++L LKE N +TI Q+YN
Sbjct: 162 PRNILLMLKEYNANGCTTIRQIYN 233
>TC219595 similar to UP|Q948R8 (Q948R8) THH1, partial (75%)
Length = 825
Score = 28.5 bits (62), Expect = 2.9
Identities = 16/48 (33%), Positives = 28/48 (58%)
Frame = -1
Query: 7 VQVDSMNNNWVPPRHMLATLKENNPGNLSTITQVYNRIKKVKELDCGP 54
V+ D +NN W+ P H+ +++ N T ++ +I+KVK+L GP
Sbjct: 423 VKQDVLNNLWLQPLHV--PIEDEN----YTPNPIHQKIEKVKDLLRGP 298
>BU545334
Length = 556
Score = 27.7 bits (60), Expect = 4.9
Identities = 12/30 (40%), Positives = 20/30 (66%)
Frame = -1
Query: 1 LTEEEKVQVDSMNNNWVPPRHMLATLKENN 30
LT++EK + + + V PR++L LKE+N
Sbjct: 94 LTKDEKTIIVDITKSMVKPRNILLILKEHN 5
>BE474062 similar to GP|9758410|dbj lysine decarboxylase-like protein
{Arabidopsis thaliana}, partial (46%)
Length = 409
Score = 26.9 bits (58), Expect = 8.3
Identities = 16/43 (37%), Positives = 21/43 (48%), Gaps = 1/43 (2%)
Frame = -2
Query: 14 NNWVPPRHMLATLKENNPGNLSTITQVYNRIKKV-KELDCGPL 55
N W PP + +K N S+I QV + K+ ELDC L
Sbjct: 288 NTWRPPSNTTCEIKPINXNTPSSINQVDVPLHKLFGELDCSVL 160
>AW757161
Length = 395
Score = 26.9 bits (58), Expect = 8.3
Identities = 15/55 (27%), Positives = 25/55 (45%), Gaps = 5/55 (9%)
Frame = -3
Query: 192 CLFH-INKNVEAKCKLWVDTTDFKALEMQKWNEVVYAETTTQFEEE----WRDMC 241
C H IN + + K+W+D L + WN ++T+ +E WR +C
Sbjct: 345 CKIHSINVLLITRKKIWLD*RQLSILSYEIWNHATIRKSTSWILKEGFPIWRVIC 181
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.321 0.135 0.419
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,361,037
Number of Sequences: 63676
Number of extensions: 170388
Number of successful extensions: 837
Number of sequences better than 10.0: 44
Number of HSP's better than 10.0 without gapping: 830
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 830
length of query: 242
length of database: 12,639,632
effective HSP length: 95
effective length of query: 147
effective length of database: 6,590,412
effective search space: 968790564
effective search space used: 968790564
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 57 (26.6 bits)
Lotus: description of TM0348.15


