

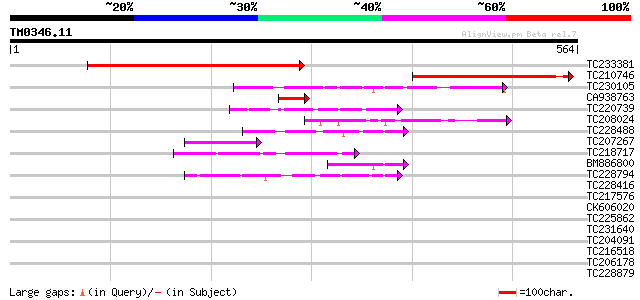
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0346.11
(564 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC233381 similar to UP|Q6NQ88 (Q6NQ88) At5g58760, partial (36%) 424 e-119
TC210746 similar to UP|Q6NQ88 (Q6NQ88) At5g58760, partial (24%) 270 2e-72
TC230105 weakly similar to UP|Q6YS10 (Q6YS10) Transducin / WD-40... 65 1e-10
CA938763 similar to GP|8843796|dbj| contains similarity to damag... 62 9e-10
TC220739 weakly similar to UP|PRP4_HUMAN (O43172) U4/U6 small nu... 50 3e-06
TC208024 49 5e-06
TC228488 similar to UP|Q8LF96 (Q8LF96) PRL1 protein, partial (70%) 47 2e-05
TC207267 similar to UP|Q8GTM1 (Q8GTM1) WD40, partial (53%) 46 4e-05
TC218717 homologue to UP|Q9M3Y1 (Q9M3Y1) Constitutively photomor... 43 3e-04
BM886800 homologue to GP|13991907|gb receptor for activated prot... 42 6e-04
TC228794 similar to UP|Q6V8V0 (Q6V8V0) Transducin-like (Fragment... 42 6e-04
TC228416 homologue to UP|MSI1_LYCES (O22466) WD-40 repeat protei... 40 0.002
TC217576 40 0.004
CK606020 39 0.005
TC225862 similar to UP|Q9W3X3 (Q9W3X3) CG3184-PA (GH14157p), par... 39 0.008
TC231640 homologue to UP|Q918P0 (Q918P0) Latency associated anti... 39 0.008
TC204091 similar to UP|Q8LLE6 (Q8LLE6) RNA-binding protein AKIP1... 39 0.008
TC216518 weakly similar to UP|Q9LW87 (Q9LW87) Coatomer protein c... 38 0.014
TC206178 similar to UP|Q8LF59 (Q8LF59) DNA-binding protein, part... 38 0.014
TC228879 similar to UP|Q6NPN9 (Q6NPN9) At1g76260, partial (30%) 37 0.031
>TC233381 similar to UP|Q6NQ88 (Q6NQ88) At5g58760, partial (36%)
Length = 653
Score = 424 bits (1091), Expect = e-119
Identities = 194/216 (89%), Positives = 206/216 (94%)
Frame = +1
Query: 78 LRKVCKVCKKPGHESGFKGATYIDCPMKPCFLCKTPGHTTLTCPHRFSTEHGVVPAPRKK 137
L+KVCKVCKKPGHE+GFKGA YIDCPMKPCFLCK PGHTTLTCPHR STEHGVVPAPR+K
Sbjct: 4 LKKVCKVCKKPGHEAGFKGAAYIDCPMKPCFLCKMPGHTTLTCPHRVSTEHGVVPAPRRK 183
Query: 138 TCKPFEYVFERQLRPGIPSIKPKYVIPDQVNCAVIRYHSRRITCLEFHPTKNNIILSGDK 197
CKP EYVFERQLRP +PSIKPKYVIPDQVNCAVIRYHSRRITCLEFHPTKNNI+LSGDK
Sbjct: 184 ACKPLEYVFERQLRPSLPSIKPKYVIPDQVNCAVIRYHSRRITCLEFHPTKNNILLSGDK 363
Query: 198 KGQLGVWDFVRVHEKVVYGNIHSCLLNNMKFNPTNDCMVYSASSDGTISFTDLETGISSS 257
KGQLGVWDF +V+EKVVYGNIHSCL+NNM+FNPTNDCMVYSASSDGTIS TDLETGISSS
Sbjct: 364 KGQLGVWDFGKVYEKVVYGNIHSCLVNNMRFNPTNDCMVYSASSDGTISCTDLETGISSS 543
Query: 258 LMNLNPEGWQGPSTWKMLYGMDINSEKGLVLVADNF 293
MNLNP+GWQGP+TWKML GMDINSEKGLVLVAD+F
Sbjct: 544 PMNLNPDGWQGPNTWKMLNGMDINSEKGLVLVADSF 651
>TC210746 similar to UP|Q6NQ88 (Q6NQ88) At5g58760, partial (24%)
Length = 872
Score = 270 bits (689), Expect = 2e-72
Identities = 135/161 (83%), Positives = 144/161 (88%)
Frame = +1
Query: 401 SREIVHSHDFNRYLTPFKAEWDPKDPSESLAVVGRYISENYNGAALHPIDFIDVSTGKLV 460
SREIVHSHDFNR+LTPFKAEWDPKDPSESLAVVGRYISENYNGAALHPIDFID STG+LV
Sbjct: 1 SREIVHSHDFNRHLTPFKAEWDPKDPSESLAVVGRYISENYNGAALHPIDFIDTSTGQLV 180
Query: 461 AEVMDPNITTISPVNKLHPNDDILASGSSSSLFIWKPKEKSEPVHEKDESKIVVCGRAEK 520
AEVMDPNITTISPVNKLHP DDILA+GSS SLFIWKPKEKSE V EKDE KIVVCG+AEK
Sbjct: 181 AEVMDPNITTISPVNKLHPRDDILATGSSRSLFIWKPKEKSELVEEKDERKIVVCGKAEK 360
Query: 521 KRGKKRGADTDESDNEGFNSKLKKSKFQKTSKSQKTKWKLS 561
KRGKK G +DESD+EGF SK +K SKS+KT+WKLS
Sbjct: 361 KRGKKNGNISDESDDEGFMSKSQK------SKSKKTEWKLS 465
>TC230105 weakly similar to UP|Q6YS10 (Q6YS10) Transducin / WD-40 repeat
protein-like, partial (23%)
Length = 939
Score = 64.7 bits (156), Expect = 1e-10
Identities = 63/279 (22%), Positives = 120/279 (42%), Gaps = 6/279 (2%)
Frame = +1
Query: 223 LNNMKFNPTNDCMVYSASSDGTISFTDLETGISSSLMNLNPEGWQGPSTWKMLYGMDI-N 281
++ P +Y++ DG D+E I + + + ++ + +
Sbjct: 49 ISGXXIQPHXXXKIYTSCYDGXXRLMDVEKEIFDLVFECD----------ESIFALSLPT 198
Query: 282 SEKGLVLVADNFGFLHLVDMRSNNKTGNAILIHKKNTKVVGIHCNPIEPDILLTCGNDHY 341
+E + +A+ +G L + D R ++ + +L H+ + +C P I+ T D
Sbjct: 199 NETNCLYLAEGYGGLTIWDNRIGKRSSHWVL-HESRINTIDFNCE--NPHIVATSSTDGT 369
Query: 342 ARVWDIRRMETGSSIYDLK---HNRVVSSAYFSPVSGNKILTTSLDNRLRVWDSIFGKMD 398
A WD+R + G + L+ H R V SAYFSP SG + TTSLDN + ++ G
Sbjct: 370 ACTWDLRYTD-GDKLRALRTFTHKRSVQSAYFSP-SGCSLATTSLDNTIGIYS---GVNL 534
Query: 399 SPSREIVHSHDFNRYLTPFKAEWDPKDPSESLAVVGRYISENYNGAALHPIDFIDVSTGK 458
+ I H++ R+L+ F+A+W D S + G +D + K
Sbjct: 535 EDAAVINHNNLTGRWLSTFRAKWGWDD------------SYLFVGNMKRGVDVVSSVERK 678
Query: 459 LVAEVMDPNITTISPVNKLHPNDDILASGSSS--SLFIW 495
+V + +++ I H + + +G++S ++IW
Sbjct: 679 IVMTLESQHMSAIPCRFDTHSYEVGMLAGATSGGQVYIW 795
>CA938763 similar to GP|8843796|dbj| contains similarity to damage-specific
DNA binding protein 2~gene_id:MZN1.20 {Arabidopsis
thaliana}, partial (5%)
Length = 446
Score = 61.6 bits (148), Expect = 9e-10
Identities = 27/31 (87%), Positives = 30/31 (96%)
Frame = +1
Query: 268 GPSTWKMLYGMDINSEKGLVLVADNFGFLHL 298
GP+TWKML GMDINSEKGLVLVAD+FGFLH+
Sbjct: 193 GPNTWKMLNGMDINSEKGLVLVADSFGFLHM 285
>TC220739 weakly similar to UP|PRP4_HUMAN (O43172) U4/U6 small nuclear
ribonucleoprotein Prp4 (U4/U6 snRNP 60 kDa protein) (WD
splicing factor Prp4) (hPrp4), partial (34%)
Length = 1206
Score = 50.1 bits (118), Expect = 3e-06
Identities = 49/174 (28%), Positives = 81/174 (46%), Gaps = 2/174 (1%)
Frame = +1
Query: 219 HSCLLNNMKFNPTNDCMVYSASSDGTISFTDLETGISSSLMNLNPEGWQGPSTWKMLYGM 278
H L + F+P+ + +AS D T D+ETG L +G S + +YG+
Sbjct: 316 HLDRLARIAFHPSGKYLG-TASFDKTWRLWDIETGDELLLQ-------EGHS--RSVYGL 465
Query: 279 DINSEKGLVLVADNFGFLHLVDMRSNNKTGNAILIHKKNTK-VVGIHCNPIEPDILLTCG 337
+++ L + D+R TG +IL + + K V+GI +P L T G
Sbjct: 466 AFHNDGSLAASCGLDSLARVWDLR----TGRSILALEGHVKPVLGISFSP-NGYHLATGG 630
Query: 338 NDHYARVWDIRRMETGSSIYDLK-HNRVVSSAYFSPVSGNKILTTSLDNRLRVW 390
D+ R+WD+R+ + S Y + H+ ++S F P G ++T S D +VW
Sbjct: 631 EDNTCRIWDLRKKK---SFYTIPAHSNLISQVKFEPHEGYFLVTASYDMTAKVW 783
>TC208024
Length = 1528
Score = 49.3 bits (116), Expect = 5e-06
Identities = 56/218 (25%), Positives = 93/218 (41%), Gaps = 12/218 (5%)
Frame = +3
Query: 294 GFLHLVDMRSNNKT-----GNAILIHKKNTKVVGIHC---NPIEPDILLTCGNDHYARVW 345
GF+ D+RSN+ T ++I +K+ VG++ +P P G+D YARV+
Sbjct: 261 GFIQHFDLRSNSATKLFCCSSSIGNNKQTLSKVGLNSIVIDPRNPYYFAIGGSDEYARVY 440
Query: 346 DIRRMETGSSIYDLKHNRVVSSAYFSP---VSGNKILTTSLDNRLRVWDSIFGKMDSPSR 402
DIR+ + GS+ +R V++ F P + N + T L + S + S +
Sbjct: 441 DIRKCQWGSA---RNSDRPVNT--FCPCHLIGSNNVHITGL-----AYSSFSELLVSYND 590
Query: 403 EIVHSHDFNRYLTPFKAEWDPKDPSESLAVVGRYISENYNGAALHPIDFIDVSTGKLVAE 462
E+++ + N + + +DP + ++ Y+G
Sbjct: 591 ELIYLFEKNVHSDSSPSSATSEDPKNI------HEAQVYSG------------------- 695
Query: 463 VMDPNITTISPVNKLHPNDDILASGSS-SSLFIWKPKE 499
N TI VN PND+ + SGS +FIWK KE
Sbjct: 696 --HRNAQTIKGVNFFGPNDEYIMSGSDCGHIFIWKKKE 803
>TC228488 similar to UP|Q8LF96 (Q8LF96) PRL1 protein, partial (70%)
Length = 1303
Score = 47.4 bits (111), Expect = 2e-05
Identities = 41/168 (24%), Positives = 75/168 (44%), Gaps = 3/168 (1%)
Frame = +3
Query: 232 NDCMVYSASSDGTISFTDLETGISSSLMNLNPEGWQGPSTWKMLYGMDINSEKGLVLVAD 291
N ++Y TI DL +G+ + + E + G+ +++ + A
Sbjct: 543 NGHVLYGMHHGRTIKIWDLASGVLKLTLTGHIE---------QVRGLAVSNRHTYMFSAG 695
Query: 292 NFGFLHLVDMRSNNKTGNAILIHKKNTKVVGIHCNPIEP--DILLTCGNDHYARVWDIRR 349
+ + D+ N +I + + G++C + P D+LLT G D RVWDIR
Sbjct: 696 DDKQVKCWDLEQNK------VIRSYHGHLSGVYCLALHPTIDVLLTGGRDSVCRVWDIR- 854
Query: 350 METGSSIYDLK-HNRVVSSAYFSPVSGNKILTTSLDNRLRVWDSIFGK 396
+ I+ L H+ V S + P + +++T S D +++WD +GK
Sbjct: 855 --SKMQIHALSGHDNTVCSVFTRP-TDPQVVTGSHDTTIKMWDLRYGK 989
>TC207267 similar to UP|Q8GTM1 (Q8GTM1) WD40, partial (53%)
Length = 1254
Score = 46.2 bits (108), Expect = 4e-05
Identities = 23/76 (30%), Positives = 37/76 (48%)
Frame = +2
Query: 175 HSRRITCLEFHPTKNNIILSGDKKGQLGVWDFVRVHEKVVYGNIHSCLLNNMKFNPTNDC 234
H+ + L++ PT++++ S G + +WD H+ +N M +N C
Sbjct: 350 HTASVEDLQWSPTESHVFASCSVDGNIAIWDTRLGKSPAASFKAHNADVNVMSWNRLASC 529
Query: 235 MVYSASSDGTISFTDL 250
M+ S S DGTIS DL
Sbjct: 530 MLASGSDDGTISIRDL 577
Score = 33.9 bits (76), Expect = 0.20
Identities = 24/112 (21%), Positives = 48/112 (42%), Gaps = 2/112 (1%)
Frame = +2
Query: 294 GFLHLVDMRSNNKTGNAILIHKKNTKVVGIHCNPIEPDILLTCGNDHYARVWDIRRMETG 353
G + + D R + H + V+ N + +L + +D + D+R ++ G
Sbjct: 422 GNIAIWDTRLGKSPAASFKAHNADVNVMS--WNRLASCMLASGSDDGTISIRDLRLLKEG 595
Query: 354 SSI--YDLKHNRVVSSAYFSPVSGNKILTTSLDNRLRVWDSIFGKMDSPSRE 403
S+ + H ++S +SP + + +S DN+L +WD K + E
Sbjct: 596 DSVVAHFEYHKHPITSIEWSPHEASSLAVSSSDNQLTIWDLSLEKDEEEEAE 751
>TC218717 homologue to UP|Q9M3Y1 (Q9M3Y1) Constitutively photomorphogenic 1
protein, partial (67%)
Length = 1880
Score = 43.1 bits (100), Expect = 3e-04
Identities = 41/187 (21%), Positives = 80/187 (41%), Gaps = 2/187 (1%)
Frame = +1
Query: 164 PDQVNCAVIRYHSR-RITCLEFHPTKNNIILSGDKKGQLGVWDFVRVHEKVVYGNIHSCL 222
P +C V+ +R +++CL ++ N I S D +G + VWD V + ++ H
Sbjct: 568 PTDAHCPVVEMSTRSKLSCLSWNKYAKNQIASSDYEGIVTVWD-VTTRKSLMEYEEHEKR 744
Query: 223 LNNMKFNPTNDCMVYSASSDGTISFTDLETGISSSLMNLNPEGWQGPSTWKMLYGMDINS 282
++ F+ T+ M+ S S D + T +S++N++ + + + N
Sbjct: 745 AWSVDFSRTDPSMLVSGSDDCKVKIWC--TNQEASVLNIDMKA--------NICCVKYNP 894
Query: 283 EKG-LVLVADNFGFLHLVDMRSNNKTGNAILIHKKNTKVVGIHCNPIEPDILLTCGNDHY 341
G + V +H D+R+ ++ + H+K V N D L + D
Sbjct: 895 GSGNYIAVGSADHHIHYYDLRNISRPVHVFSGHRKAVSYVKFLSN----DELASASTDST 1062
Query: 342 ARVWDIR 348
R+WD++
Sbjct: 1063LRLWDVK 1083
>BM886800 homologue to GP|13991907|gb receptor for activated protein kinase C
RACK1 {Heliothis virescens}, partial (43%)
Length = 428
Score = 42.4 bits (98), Expect = 6e-04
Identities = 28/83 (33%), Positives = 38/83 (45%), Gaps = 3/83 (3%)
Frame = +1
Query: 317 NTKVVGIHCNPIEPDILLTCGNDHYARVWDIRRMETGSSIYDLK---HNRVVSSAYFSPV 373
N V I NP PD++L+C D VW + R + I + H+ VS S
Sbjct: 43 NGWVTQIATNPKYPDMILSCSRDKTLIVWKLTRDDHSYGIPQKRLYGHSHFVSDVVLSS- 219
Query: 374 SGNKILTTSLDNRLRVWDSIFGK 396
GN L+ S D LR+WD G+
Sbjct: 220 DGNYALSGSWDKTLRLWDLAAGR 288
>TC228794 similar to UP|Q6V8V0 (Q6V8V0) Transducin-like (Fragment), complete
Length = 870
Score = 42.4 bits (98), Expect = 6e-04
Identities = 47/218 (21%), Positives = 94/218 (42%), Gaps = 2/218 (0%)
Frame = +3
Query: 175 HSRRITCLEFHPTKNNIILSGDKKGQLGVWDFVRVHEKVVYGNIHSCLLNNMKFNPTNDC 234
H I ++F+P +++ SG ++ +W+ + + H + ++ + T+
Sbjct: 276 HQSAIYTMKFNPA-GSVVASGSHDREIFLWNVHGDCKNFMVLKGHKNAVLDLHWT-TDGT 449
Query: 235 MVYSASSDGTISFTDLETG--ISSSLMNLNPEGWQGPSTWKMLYGMDINSEKGLVLVADN 292
+ SAS D T+ D+ETG I + +L+ PS LV+ +
Sbjct: 450 QIVSASPDKTVRAWDVETGKQIKKMVEHLSYVNSCCPS----------RRGPPLVVSGSD 599
Query: 293 FGFLHLVDMRSNNKTGNAILIHKKNTKVVGIHCNPIEPDILLTCGNDHYARVWDIRRMET 352
G L DMR +I ++ + + D + T G D+ ++WD+R+ E
Sbjct: 600 DGTAKLWDMRQR----GSIQTFPDKYQITAVGFSDAS-DKIFTGGIDNDVKIWDLRKGEV 764
Query: 353 GSSIYDLKHNRVVSSAYFSPVSGNKILTTSLDNRLRVW 390
++ H ++++ SP G+ +LT +D +L +W
Sbjct: 765 TMTLQG--HQDMITAMQLSP-DGSYLLTNGMDCKLCIW 869
Score = 34.7 bits (78), Expect = 0.12
Identities = 18/62 (29%), Positives = 35/62 (56%)
Frame = +3
Query: 330 PDILLTCGNDHYARVWDIRRMETGSSIYDLKHNRVVSSAYFSPVSGNKILTTSLDNRLRV 389
P ++++ +D A++WD+R+ SI +++ FS S +KI T +DN +++
Sbjct: 573 PPLVVSGSDDGTAKLWDMRQR---GSIQTFPDKYQITAVGFSDAS-DKIFTGGIDNDVKI 740
Query: 390 WD 391
WD
Sbjct: 741 WD 746
>TC228416 homologue to UP|MSI1_LYCES (O22466) WD-40 repeat protein MSI1,
partial (86%)
Length = 1416
Score = 40.4 bits (93), Expect = 0.002
Identities = 56/277 (20%), Positives = 102/277 (36%), Gaps = 23/277 (8%)
Frame = +3
Query: 137 KTCKPFEYVFERQLRPGIPSIKPKYVIPDQVNCAVIRYHSRRITCLEFHPTKNNIILSGD 196
KT YVF+ P P + + PD +R H+ L + K +LSG
Sbjct: 282 KTVSAEVYVFDYSKHPSKPPLDG-FCNPD----LRLRGHNTEGYGLSWSKFKQGHLLSGS 446
Query: 197 KKGQLGVWDF-----VRVHEKVVYGNIHSCLLNNMKFNPTNDCMVYSASSDGTISFTDLE 251
Q+ +WD + E + +H ++ ++ ++ ++ + S D + DL
Sbjct: 447 DDAQICLWDINGTPKNKSLEAMQIFKVHEGVVEDVAWHLRHEYLFGSVGDDQYLLIWDLR 626
Query: 252 TGISSSLMNLNPEGWQGPSTWKMLYGMDINS------EKGLVLVADNFGFLHLVDMRSNN 305
T +S P + + ++N + +V + L D+R N
Sbjct: 627 TPAASK-----------PVQSVVAHQSEVNCLAFNPFNEWVVATGSTDKTVKLFDLRKIN 773
Query: 306 KTGNAILIHKKNTKVVGIHCNPIEPDILLTCGNDHYARVWDIRRMETGSSIYDLK----- 360
+ HK+ VG NP IL +C VWD+ R++ S D +
Sbjct: 774 TPLHIFDSHKEEVFQVG--WNPKNETILASCCLGRRLMVWDLSRIDEEQSPEDAEDGPPE 947
Query: 361 -------HNRVVSSAYFSPVSGNKILTTSLDNRLRVW 390
H +S ++P + + + DN L++W
Sbjct: 948 LLFIHGGHTSKISDFSWNPCEDWVVASVAEDNILQIW 1058
Score = 35.8 bits (81), Expect = 0.052
Identities = 23/99 (23%), Positives = 47/99 (47%), Gaps = 2/99 (2%)
Frame = +3
Query: 295 FLHLVDMRSN--NKTGNAILIHKKNTKVVGIHCNPIEPDILLTCGNDHYARVWDIRRMET 352
+L + D+R+ +K +++ H+ + NP ++ T D +++D+R++ T
Sbjct: 603 YLLIWDLRTPAASKPVQSVVAHQSEVNCLAF--NPFNEWVVATGSTDKTVKLFDLRKINT 776
Query: 353 GSSIYDLKHNRVVSSAYFSPVSGNKILTTSLDNRLRVWD 391
I+D H V ++P + + + L RL VWD
Sbjct: 777 PLHIFD-SHKEEVFQVGWNPKNETILASCCLGRRLMVWD 890
Score = 34.7 bits (78), Expect = 0.12
Identities = 30/123 (24%), Positives = 58/123 (46%), Gaps = 2/123 (1%)
Frame = +3
Query: 305 NKTGNAILIHKKNTKVVGIHCNPIEPDILL-TCGNDHYARVWDIRRMETGSSIYD-LKHN 362
NK+ A+ I K + VV + + L + G+D Y +WD+R + + H
Sbjct: 492 NKSLEAMQIFKVHEGVVEDVAWHLRHEYLFGSVGDDQYLLIWDLRTPAASKPVQSVVAHQ 671
Query: 363 RVVSSAYFSPVSGNKILTTSLDNRLRVWDSIFGKMDSPSREIVHSHDFNRYLTPFKAEWD 422
V+ F+P + + T S D ++++D K+++P +H D ++ F+ W+
Sbjct: 672 SEVNCLAFNPFNEWVVATGSTDKTVKLFD--LRKINTP----LHIFDSHKE-EVFQVGWN 830
Query: 423 PKD 425
PK+
Sbjct: 831 PKN 839
>TC217576
Length = 1432
Score = 39.7 bits (91), Expect = 0.004
Identities = 24/86 (27%), Positives = 43/86 (49%), Gaps = 3/86 (3%)
Frame = +1
Query: 323 IHCNPI-EPDILLTCGNDHYARVWDIRRMETGSSIYDLKHNRVVSSAYFSPVSGNKILTT 381
+H PI +P LL+ D R+W + + HN V+ F+PV+ N ++
Sbjct: 88 LHQAPILKPQFLLSSSVDKTVRLWHVG---IDRCLRVFSHNNYVTCVNFNPVNDNFFISG 258
Query: 382 SLDNRLRVWDSIFGKMDS--PSREIV 405
S+D ++R+W+ + ++ REIV
Sbjct: 259 SIDGKVRIWEVVHCRVSDYIDIREIV 336
Score = 38.9 bits (89), Expect = 0.006
Identities = 16/65 (24%), Positives = 33/65 (50%), Gaps = 12/65 (18%)
Frame = +1
Query: 155 PSIKPKYVIPDQVN------------CAVIRYHSRRITCLEFHPTKNNIILSGDKKGQLG 202
P +KP++++ V+ C + H+ +TC+ F+P +N +SG G++
Sbjct: 100 PILKPQFLLSSSVDKTVRLWHVGIDRCLRVFSHNNYVTCVNFNPVNDNFFISGSIDGKVR 279
Query: 203 VWDFV 207
+W+ V
Sbjct: 280 IWEVV 294
>CK606020
Length = 340
Score = 39.3 bits (90), Expect = 0.005
Identities = 19/42 (45%), Positives = 27/42 (64%)
Frame = +1
Query: 20 DSEQSSSEEEEEALEDDEEEALEHEQEEENGVAEIGKNEKLE 61
+ E+ EEEEE E++EEE E E+EEE V E + E++E
Sbjct: 199 EEEEEEEEEEEEEEEEEEEEIEEEEEEEEEEVEEEVEEEEVE 324
Score = 39.3 bits (90), Expect = 0.005
Identities = 18/39 (46%), Positives = 24/39 (61%)
Frame = +1
Query: 15 IERDTDSEQSSSEEEEEALEDDEEEALEHEQEEENGVAE 53
I+ D E+ EEEEE E++EEE +E E+EEE E
Sbjct: 181 IDPSLDEEEEEEEEEEEEEEEEEEEEIEEEEEEEEEEVE 297
Score = 35.0 bits (79), Expect = 0.089
Identities = 19/51 (37%), Positives = 28/51 (54%)
Frame = +1
Query: 11 PKVVIERDTDSEQSSSEEEEEALEDDEEEALEHEQEEENGVAEIGKNEKLE 61
P + E + + E+ EEEEE E +EEE E E+ EE E + E++E
Sbjct: 187 PSLDEEEEEEEEEEEEEEEEEEEEIEEEEEEEEEEVEEEVEEEEVEGEEVE 339
Score = 33.9 bits (76), Expect = 0.20
Identities = 20/54 (37%), Positives = 27/54 (49%)
Frame = +1
Query: 8 VPFPKVVIERDTDSEQSSSEEEEEALEDDEEEALEHEQEEENGVAEIGKNEKLE 61
+P V + + D EEEEE E++EEE E E EEE E E++E
Sbjct: 148 LPDSPVANDDEIDPSLDEEEEEEEEEEEEEEEEEEEEIEEEEEEEEEEVEEEVE 309
Score = 33.5 bits (75), Expect = 0.26
Identities = 20/52 (38%), Positives = 28/52 (53%)
Frame = +1
Query: 11 PKVVIERDTDSEQSSSEEEEEALEDDEEEALEHEQEEENGVAEIGKNEKLEV 62
P + D + + S EEEEE +EEE E E+EEE + E + E+ EV
Sbjct: 151 PDSPVANDDEIDPSLDEEEEE----EEEEEEEEEEEEEEEIEEEEEEEEEEV 294
>TC225862 similar to UP|Q9W3X3 (Q9W3X3) CG3184-PA (GH14157p), partial (14%)
Length = 862
Score = 38.5 bits (88), Expect = 0.008
Identities = 44/201 (21%), Positives = 80/201 (38%), Gaps = 12/201 (5%)
Frame = +2
Query: 211 EKVVYGNIHSCLLNNMKFNPTNDCMVYSASSDGTISFTDLETGISSSLMNLNPEGWQGPS 270
+++ G I + + + +NP+ + S DG++S L S + + E W+
Sbjct: 74 KEITNGKISNSMCLYLDWNPSATSITVGLS-DGSVSIVSL----LESKLEIQDE-WKAHD 235
Query: 271 TWKMLYGMDINSEKGLVLVADNFGFLHLVDMRSNNKTGNAILIHKKNTKVVGIHC---NP 327
DI+ + +D+ F D+R +K N + K K+ G+ C +P
Sbjct: 236 FELWTTSFDIHQPNLVYTGSDDCKF-SCWDLR--DKPPNVVFQSSKVHKM-GVCCIEKSP 403
Query: 328 IEPDILLTCGNDHYARVWDIRRM---------ETGSSIYDLKHNRVVSSAYFSPVSGNKI 378
+P+ LLT D Y RVWD+R M G ++ +KH+ + + N
Sbjct: 404 HDPNTLLTGSYDEYLRVWDLRSMSKPLNKTSINLGGGVWRVKHHPFIPGLVLAACMHNGF 583
Query: 379 LTTSLDNRLRVWDSIFGKMDS 399
++ + K DS
Sbjct: 584 AIVAIKGNNAEVSETYKKHDS 646
>TC231640 homologue to UP|Q918P0 (Q918P0) Latency associated antigen,
partial (7%)
Length = 466
Score = 38.5 bits (88), Expect = 0.008
Identities = 19/44 (43%), Positives = 25/44 (56%)
Frame = +1
Query: 16 ERDTDSEQSSSEEEEEALEDDEEEALEHEQEEENGVAEIGKNEK 59
E + E+ EEEEE E++EEE E E+EEE V I +K
Sbjct: 34 EEKEEEEEEEEEEEEEEEEEEEEEEKEEEEEEEGNVLVIIDEDK 165
Score = 37.7 bits (86), Expect = 0.014
Identities = 17/31 (54%), Positives = 21/31 (66%)
Frame = +1
Query: 18 DTDSEQSSSEEEEEALEDDEEEALEHEQEEE 48
DT E+ EEEEE E++EEE E E+EEE
Sbjct: 16 DTQEEEEEKEEEEEEEEEEEEEEEEEEEEEE 108
Score = 37.4 bits (85), Expect = 0.018
Identities = 16/35 (45%), Positives = 23/35 (65%)
Frame = +1
Query: 16 ERDTDSEQSSSEEEEEALEDDEEEALEHEQEEENG 50
E + + E+ EEEEE E++EEE + E+EEE G
Sbjct: 31 EEEKEEEEEEEEEEEEEEEEEEEEEEKEEEEEEEG 135
Score = 36.2 bits (82), Expect = 0.040
Identities = 19/42 (45%), Positives = 26/42 (61%)
Frame = +1
Query: 20 DSEQSSSEEEEEALEDDEEEALEHEQEEENGVAEIGKNEKLE 61
D+ S ++EEEE E++EEE E E+EEE E K E+ E
Sbjct: 1 DNVLSDTQEEEEEKEEEEEEEEEEEEEEEEEEEEEEKEEEEE 126
Score = 35.0 bits (79), Expect = 0.089
Identities = 17/43 (39%), Positives = 25/43 (57%)
Frame = +1
Query: 13 VVIERDTDSEQSSSEEEEEALEDDEEEALEHEQEEENGVAEIG 55
V+ + + E+ EEEEE E++EEE E E+E+E E G
Sbjct: 7 VLSDTQEEEEEKEEEEEEEEEEEEEEEEEEEEEEKEEEEEEEG 135
Score = 34.3 bits (77), Expect = 0.15
Identities = 18/39 (46%), Positives = 24/39 (61%)
Frame = +1
Query: 25 SSEEEEEALEDDEEEALEHEQEEENGVAEIGKNEKLEVG 63
+ EEEEE E++EEE E E+EEE E + E+ E G
Sbjct: 19 TQEEEEEKEEEEEEEEEEEEEEEEEEEEEEKEEEEEEEG 135
Score = 31.6 bits (70), Expect = 0.99
Identities = 17/44 (38%), Positives = 24/44 (53%)
Frame = +1
Query: 18 DTDSEQSSSEEEEEALEDDEEEALEHEQEEENGVAEIGKNEKLE 61
D + EEEE+ E++EEE E E+EEE E + E+ E
Sbjct: 1 DNVLSDTQEEEEEKEEEEEEEEEEEEEEEEEEEEEEKEEEEEEE 132
>TC204091 similar to UP|Q8LLE6 (Q8LLE6) RNA-binding protein AKIP1, partial
(70%)
Length = 2075
Score = 38.5 bits (88), Expect = 0.008
Identities = 18/36 (50%), Positives = 24/36 (66%)
Frame = +2
Query: 18 DTDSEQSSSEEEEEALEDDEEEALEHEQEEENGVAE 53
+ + E+ EEEEE ED+EEE E E+EEEN V +
Sbjct: 152 EEEVEEEVEEEEEEEEEDEEEEEEEEEEEEENDVVK 259
Score = 35.0 bits (79), Expect = 0.089
Identities = 15/45 (33%), Positives = 27/45 (59%)
Frame = +2
Query: 15 IERDTDSEQSSSEEEEEALEDDEEEALEHEQEEENGVAEIGKNEK 59
+E + + E+ E EEE E++E+E E E+EEE ++ K ++
Sbjct: 134 VEEEVEEEEVEEEVEEEEEEEEEDEEEEEEEEEEEEENDVVKQQQ 268
Score = 34.3 bits (77), Expect = 0.15
Identities = 17/39 (43%), Positives = 24/39 (60%)
Frame = +2
Query: 10 FPKVVIERDTDSEQSSSEEEEEALEDDEEEALEHEQEEE 48
+ +V E + + + EEEEE E+DEEE E E+EEE
Sbjct: 125 YEEVEEEVEEEEVEEEVEEEEEEEEEDEEEEEEEEEEEE 241
Score = 34.3 bits (77), Expect = 0.15
Identities = 20/57 (35%), Positives = 33/57 (57%), Gaps = 1/57 (1%)
Frame = +2
Query: 3 PVTRRVPFPKVVIERDTDSEQSSSEEE-EEALEDDEEEALEHEQEEENGVAEIGKNE 58
P+ ++ P P+ + + E+ EEE EE +E++EEE E E+EEE E +N+
Sbjct: 80 PLKQQQPKPENPPQEYEEVEEEVEEEEVEEEVEEEEEEEEEDEEEEEEEEEEEEEND 250
>TC216518 weakly similar to UP|Q9LW87 (Q9LW87) Coatomer protein complex, beta
prime; beta'-COP protein, partial (29%)
Length = 1189
Score = 37.7 bits (86), Expect = 0.014
Identities = 44/174 (25%), Positives = 70/174 (39%), Gaps = 2/174 (1%)
Frame = +1
Query: 175 HSRRITCLEFHPTKNNIILSGDKKGQLGVWDFVRVHEKVVYGNIHSCLLNNMKFNPTNDC 234
H I L HP +I + D + L +W++ + HS + + FNP +
Sbjct: 343 HKDYIRSLAVHPVLPYVISASDDQ-VLKLWNWRKGWSCYENFEGHSHYVMQVAFNPKDPS 519
Query: 235 MVYSASSDGTISFTDLETGISSSLMNLNPEGWQGPSTWKMLYGMD--INSEKGLVLVADN 292
SAS DGT+ L+ SS N EG Q K + +D I ++K +L +
Sbjct: 520 TFASASLDGTLKIWSLD----SSAPNFTLEGHQ-----KGVNCVDYFITNDKQYLLSGSD 672
Query: 293 FGFLHLVDMRSNNKTGNAILIHKKNTKVVGIHCNPIEPDILLTCGNDHYARVWD 346
+ D S N + H+ N + C E I++T D ++WD
Sbjct: 673 DYTAKVWDYHSRNCV-QTLEGHENNVTAI---CAHPELPIIITASEDSTVKIWD 822
Score = 32.3 bits (72), Expect = 0.58
Identities = 20/79 (25%), Positives = 39/79 (49%), Gaps = 2/79 (2%)
Frame = +1
Query: 314 HKKNTKVVGIHCNPIEPDILLTCGNDHYARVWDIRRMETGSSIYDL--KHNRVVSSAYFS 371
HK + + +H P+ P ++ + +D ++W+ R+ G S Y+ H+ V F+
Sbjct: 343 HKDYIRSLAVH--PVLPYVI-SASDDQVLKLWNWRK---GWSCYENFEGHSHYVMQVAFN 504
Query: 372 PVSGNKILTTSLDNRLRVW 390
P + + SLD L++W
Sbjct: 505 PKDPSTFASASLDGTLKIW 561
>TC206178 similar to UP|Q8LF59 (Q8LF59) DNA-binding protein, partial (69%)
Length = 1138
Score = 37.7 bits (86), Expect = 0.014
Identities = 19/55 (34%), Positives = 26/55 (46%), Gaps = 13/55 (23%)
Frame = +2
Query: 81 VCKVCKKPGHES------------GFKGATYIDCPMKP-CFLCKTPGHTTLTCPH 122
+CK CK+PGH + G G +C K C+ CK PGH +CP+
Sbjct: 341 LCKNCKRPGHYARECPNVAICHNCGLPGHIASECTTKSLCWNCKEPGHMASSCPN 505
Score = 34.3 bits (77), Expect = 0.15
Identities = 17/43 (39%), Positives = 19/43 (43%), Gaps = 1/43 (2%)
Frame = +2
Query: 80 KVCKVCKKPGHESGFKGATYIDCPMKP-CFLCKTPGHTTLTCP 121
K C C+K GH + DCP P C LC GH CP
Sbjct: 644 KACNNCRKTGHLAR-------DCPNDPICNLCNVSGHVARQCP 751
>TC228879 similar to UP|Q6NPN9 (Q6NPN9) At1g76260, partial (30%)
Length = 742
Score = 36.6 bits (83), Expect = 0.031
Identities = 16/68 (23%), Positives = 35/68 (50%)
Frame = +3
Query: 179 ITCLEFHPTKNNIILSGDKKGQLGVWDFVRVHEKVVYGNIHSCLLNNMKFNPTNDCMVYS 238
+ ++FHP K ++++ + + + +WD + + H+ +K NP D M+ S
Sbjct: 42 VXSVDFHPQKQXMLVTAEHESGIHIWDLRKPKVPIQELPGHTHWTWTVKCNPEYDGMILS 221
Query: 239 ASSDGTIS 246
A +D T++
Sbjct: 222 AGTDSTVN 245
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.315 0.134 0.404
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 27,729,596
Number of Sequences: 63676
Number of extensions: 446125
Number of successful extensions: 2664
Number of sequences better than 10.0: 150
Number of HSP's better than 10.0 without gapping: 2061
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2389
length of query: 564
length of database: 12,639,632
effective HSP length: 102
effective length of query: 462
effective length of database: 6,144,680
effective search space: 2838842160
effective search space used: 2838842160
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0346.11


