

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
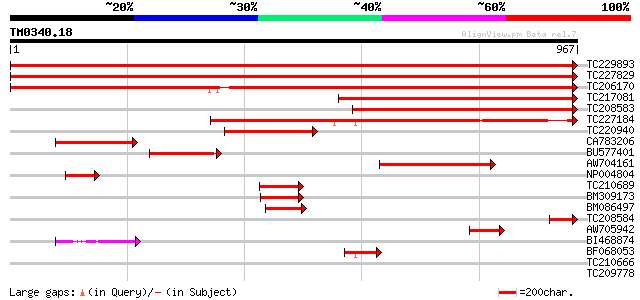
Query= TM0340.18
(967 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC229893 UP|Q8H928 (Q8H928) Phosphoenolpyruvate carboxylase , c... 1773 0.0
TC227829 UP|CAP2_SOYBN (P51061) Phosphoenolpyruvate carboxylase ... 1772 0.0
TC206170 UP|CAP1_SOYBN (Q02909) Phosphoenolpyruvate carboxylase,... 1754 0.0
TC217081 homologue to UP|Q6Q2Z8 (Q6Q2Z8) Phosphoenolpyruvate car... 756 0.0
TC208583 Phosphoenolpyruvate carboxylase [Glycine max] 702 0.0
TC227184 UP|Q6Q2Z9 (Q6Q2Z9) Phosphoenolpyruvate carboxylase , p... 544 e-155
TC220940 homologue to UP|Q9T0N1 (Q9T0N1) Ppc2 protein , partial... 275 5e-74
CA783206 similar to PIR|T09846|T09 phosphoenolpyruvate carboxyla... 244 2e-64
BU577401 homologue to EGAD|139142|14 ppc2 protein {Mesembryanthe... 226 3e-59
AW704161 weakly similar to GP|18073660|em phosphoenolpyruvate ca... 221 1e-57
NP004804 phosphoenolpyruvate carboxylase 96 8e-20
TC210689 homologue to UP|Q6Q2Z9 (Q6Q2Z9) Phosphoenolpyruvate car... 92 1e-18
BM309173 90 5e-18
BM086497 82 1e-15
TC208584 homologue to UP|O22119 (O22119) Phosphoenolpyruvate car... 71 2e-12
AW705942 weakly similar to GP|25901015|dbj phosphoenolpyruvate c... 69 1e-11
BI468874 56 9e-08
BF068053 50 6e-06
TC210666 32 1.8
TC209778 similar to UP|Q88GI3 (Q88GI3) Major facilitator family ... 27 2.2
>TC229893 UP|Q8H928 (Q8H928) Phosphoenolpyruvate carboxylase , complete
Length = 3536
Score = 1773 bits (4593), Expect = 0.0
Identities = 878/967 (90%), Positives = 921/967 (94%)
Frame = +3
Query: 1 MANRNLEKMASIDAQLRQLVPAKVSEDDKLVEYDALLLDRFLDILQDLHGEDLKETVQEV 60
MANRNLEKMASIDAQLRQL PAKVSEDDKL+EYDALLLDRFLDILQDLHGEDLKETVQEV
Sbjct: 204 MANRNLEKMASIDAQLRQLAPAKVSEDDKLIEYDALLLDRFLDILQDLHGEDLKETVQEV 383
Query: 61 YELSAEYERKHEPQRLEVLGNLITSLDAGDSIVVAKSFAHMLNLANLAEEVQIAHRRRIK 120
YELSAEYE KH+P++LE LGNLITSLDAGDSI+VAKSF+HMLNLANLAEEVQI+ RRR K
Sbjct: 384 YELSAEYEGKHDPKKLEELGNLITSLDAGDSILVAKSFSHMLNLANLAEEVQISRRRRNK 563
Query: 121 LKKGDFADEGNATTESDIEETFKKLVGEMKKSPQEVFDELKNQTVDLVLTAHPTQSVRRS 180
LKKGDFADE NATTESDIEET KKLV +KKSPQEVFD LKNQTVDLVLTAHPTQS+RRS
Sbjct: 564 LKKGDFADENNATTESDIEETLKKLVFGLKKSPQEVFDALKNQTVDLVLTAHPTQSIRRS 743
Query: 181 LLQKHGRIRNCLTQLYAKDITPDDKQELDEALQREIQAAFRTDEIRRTAPTPQDEMRAGM 240
LLQKHGRIRNCL+QLYAKDITPDDKQELDEALQREIQAAFRTDEIRRT PTPQDEMRAGM
Sbjct: 744 LLQKHGRIRNCLSQLYAKDITPDDKQELDEALQREIQAAFRTDEIRRTPPTPQDEMRAGM 923
Query: 241 SYFHETIWKGVPKFLRRVDTALKNIGINERVPYNAPLIQFSSWMGGDRDGNPRVTPEVTR 300
SYFHETIW GVP+FLRRVDTAL NIGI ERVPYNAPLIQFSSWMGGDRDGNPRVTPEVTR
Sbjct: 924 SYFHETIWNGVPRFLRRVDTALNNIGIKERVPYNAPLIQFSSWMGGDRDGNPRVTPEVTR 1103
Query: 301 DVCLLARMMAANLYYSQIEDLMFELSMWRCNDELRVRAEELTSSSNKDAVAKHYIEFWKN 360
DVCLLARMMAANLYYSQIEDLMFELSMWRCNDELRVRAEEL SS KD VAKHYIEFWK
Sbjct: 1104 DVCLLARMMAANLYYSQIEDLMFELSMWRCNDELRVRAEELHRSSKKDEVAKHYIEFWKK 1283
Query: 361 IPPSEPYRVVLGEVRNRLYQTRERSRHLLANEYSDILEEHTFTNVEEFLEPLELCYRSLC 420
+PP+EPYRVVLGEVR+RLYQTRERSRHLL+N YSDI EE TFTNVEEFLE LELCYRSLC
Sbjct: 1284 VPPNEPYRVVLGEVRDRLYQTRERSRHLLSNGYSDIPEEATFTNVEEFLESLELCYRSLC 1463
Query: 421 ACGDRAIADGSLLDFLRQVSTFGLSLVRLDIRQESDRHTDVLDAITKHLGIGSYLEWSEE 480
ACGDRAIADGSLLDF+RQVSTFGLSLVRLDIRQESDRHTDVLDAITKHL IGSY EWSEE
Sbjct: 1464 ACGDRAIADGSLLDFMRQVSTFGLSLVRLDIRQESDRHTDVLDAITKHLEIGSYQEWSEE 1643
Query: 481 KRQQWLLSELSGKRPLFGPDLPQTEEIKDVLDTFHVIAELPSDNFGAYIISMATAPSDVL 540
KRQ+WLLSELSGKRPLFGPDLPQTEEI+DVLDTFHVIAELP DNFGAYIISMATAPSDVL
Sbjct: 1644 KRQEWLLSELSGKRPLFGPDLPQTEEIRDVLDTFHVIAELPPDNFGAYIISMATAPSDVL 1823
Query: 541 AVELLQRECHVKNPLRVVPLFEKLADLETAPAALARLFSVEWYRNRINGKQEVMIGYSDS 600
AVELLQRECH+K+PLRVVPLFEKLADLE APAALARLFS++WYRNRINGKQEVMIGYSDS
Sbjct: 1824 AVELLQRECHIKHPLRVVPLFEKLADLEAAPAALARLFSIDWYRNRINGKQEVMIGYSDS 2003
Query: 601 GKDAGRFSAAWQLYKAQEELIKVAKEYGVKLTMFHGRGGTVGRGGGPTHLAILSQPPDTI 660
GKDAGRFSAAWQLYKAQEELI VAK++G+KLTMFHGRGGTVGRGGGPTHLAILSQPPDTI
Sbjct: 2004 GKDAGRFSAAWQLYKAQEELINVAKKFGIKLTMFHGRGGTVGRGGGPTHLAILSQPPDTI 2183
Query: 661 HGSLRVTVQGEVIEQSFGEEHLCFRTLQRYTAATLEHGMHPPISPKPEWRALMDEMAVIA 720
HGSLRVTVQGEVIEQSFGE+HLCFRTLQR+TAATLEHGMHPPISPKPEWRALMD+MAVIA
Sbjct: 2184 HGSLRVTVQGEVIEQSFGEQHLCFRTLQRFTAATLEHGMHPPISPKPEWRALMDQMAVIA 2363
Query: 721 TEEYRSIVFKEPRFVEYFRLATPELEYGRMNIGSRPAKRKPSGGIETLRAIPWIFAWTQT 780
TEEYRSIVFKEPRFVEYFRLATPELEYGRMNIGSRPAKRKPSGGIETLRAIPWIFAWTQT
Sbjct: 2364 TEEYRSIVFKEPRFVEYFRLATPELEYGRMNIGSRPAKRKPSGGIETLRAIPWIFAWTQT 2543
Query: 781 RFHLPVWLGFGAAFKHVIAKDIRNLNMLQEMYNQWPFFRVTIDLVEMVFAKGDPGIAALY 840
RFHLPVWLGFGAAFK VI K++ NLNMLQEMYNQWPFFRVT+DLVEMVFAKGDP IA L
Sbjct: 2544 RFHLPVWLGFGAAFKEVIEKNVNNLNMLQEMYNQWPFFRVTLDLVEMVFAKGDPKIAGLN 2723
Query: 841 DRLLVSEDLWSFGEQLRTMFEETKQLLLQVAAHKDLLEGDPYLKQRLRLRDSYITTLNVC 900
DRLLVS+DLW FG+QLR +EETK+LLLQVA HK++LEGDPYLKQRLRLR + ITTLN+
Sbjct: 2724 DRLLVSKDLWLFGDQLRNKYEETKKLLLQVAGHKEILEGDPYLKQRLRLRHAPITTLNIV 2903
Query: 901 QAYTLKRIRDPNYNVKLRPHISKEAIDVSKPADELVTLNPTSEYAPGLEDTLILTMKGIA 960
QAYTLKRIRDPNYNVK+RP ISKE+ + SK ADEL+ LNPTSEYAPGLEDTLILTMKGIA
Sbjct: 2904 QAYTLKRIRDPNYNVKVRPRISKESAEASKSADELIKLNPTSEYAPGLEDTLILTMKGIA 3083
Query: 961 AGMQNTG 967
AGMQNTG
Sbjct: 3084 AGMQNTG 3104
>TC227829 UP|CAP2_SOYBN (P51061) Phosphoenolpyruvate carboxylase (PEPCase) ,
complete
Length = 3192
Score = 1772 bits (4589), Expect = 0.0
Identities = 877/967 (90%), Positives = 923/967 (94%)
Frame = +1
Query: 1 MANRNLEKMASIDAQLRQLVPAKVSEDDKLVEYDALLLDRFLDILQDLHGEDLKETVQEV 60
MA RNLEKMASIDAQLRQL PAKVSEDDKL+EYDALLLDRFLDILQDLHGEDLKETVQEV
Sbjct: 43 MATRNLEKMASIDAQLRQLAPAKVSEDDKLIEYDALLLDRFLDILQDLHGEDLKETVQEV 222
Query: 61 YELSAEYERKHEPQRLEVLGNLITSLDAGDSIVVAKSFAHMLNLANLAEEVQIAHRRRIK 120
YELSAEYE KH+P++LE LGNLITSLDAGDSI+VAKSF+HMLNLANLAEEVQI+ RRR K
Sbjct: 223 YELSAEYEGKHDPKKLEELGNLITSLDAGDSILVAKSFSHMLNLANLAEEVQISRRRRNK 402
Query: 121 LKKGDFADEGNATTESDIEETFKKLVGEMKKSPQEVFDELKNQTVDLVLTAHPTQSVRRS 180
LKKGDFADE NATTESDIEET KKLV ++KKSPQEVFD LKNQTVDLVLTAHPTQS+RRS
Sbjct: 403 LKKGDFADENNATTESDIEETLKKLVFDLKKSPQEVFDALKNQTVDLVLTAHPTQSIRRS 582
Query: 181 LLQKHGRIRNCLTQLYAKDITPDDKQELDEALQREIQAAFRTDEIRRTAPTPQDEMRAGM 240
LLQKHGRIRNCL+QLYAKDITPDDKQELDEALQREIQAAFRTDEIRRT PTPQDEMRAGM
Sbjct: 583 LLQKHGRIRNCLSQLYAKDITPDDKQELDEALQREIQAAFRTDEIRRTPPTPQDEMRAGM 762
Query: 241 SYFHETIWKGVPKFLRRVDTALKNIGINERVPYNAPLIQFSSWMGGDRDGNPRVTPEVTR 300
SYFHETIW GVP+FLRRVDTAL NIGI ERVPYNAPLIQFSSWMGGDRDGNPRVTPEVTR
Sbjct: 763 SYFHETIWNGVPRFLRRVDTALNNIGIKERVPYNAPLIQFSSWMGGDRDGNPRVTPEVTR 942
Query: 301 DVCLLARMMAANLYYSQIEDLMFELSMWRCNDELRVRAEELTSSSNKDAVAKHYIEFWKN 360
DVCLLARMMAANLYYSQIEDLMFELSMWRCNDELRVRAEEL SS KD VAKHYIEFWK
Sbjct: 943 DVCLLARMMAANLYYSQIEDLMFELSMWRCNDELRVRAEELHRSSKKDEVAKHYIEFWKK 1122
Query: 361 IPPSEPYRVVLGEVRNRLYQTRERSRHLLANEYSDILEEHTFTNVEEFLEPLELCYRSLC 420
+PP+EPYRVVLGEVR+RLYQTRERSRHLL+N YSDI EE TFTNVEEFLE LELCYRSLC
Sbjct: 1123 VPPNEPYRVVLGEVRDRLYQTRERSRHLLSNGYSDIPEEATFTNVEEFLESLELCYRSLC 1302
Query: 421 ACGDRAIADGSLLDFLRQVSTFGLSLVRLDIRQESDRHTDVLDAITKHLGIGSYLEWSEE 480
ACGDRAIADGSLLDF+RQVSTFGLSLVRLDIRQESDRHTDVLDAITKHL IGSY EWSEE
Sbjct: 1303 ACGDRAIADGSLLDFMRQVSTFGLSLVRLDIRQESDRHTDVLDAITKHLEIGSYQEWSEE 1482
Query: 481 KRQQWLLSELSGKRPLFGPDLPQTEEIKDVLDTFHVIAELPSDNFGAYIISMATAPSDVL 540
KRQ+WLLSELSGKRPLFGPDLPQTEEI+DVLDTFHVIAELP DNFGAYIISMATAPSDVL
Sbjct: 1483 KRQEWLLSELSGKRPLFGPDLPQTEEIRDVLDTFHVIAELPPDNFGAYIISMATAPSDVL 1662
Query: 541 AVELLQRECHVKNPLRVVPLFEKLADLETAPAALARLFSVEWYRNRINGKQEVMIGYSDS 600
AVELLQRECH+K+PLRVVPLFEKLADLE APAALARLFS++WYRNRINGKQEVMIGYSDS
Sbjct: 1663 AVELLQRECHIKHPLRVVPLFEKLADLEAAPAALARLFSIDWYRNRINGKQEVMIGYSDS 1842
Query: 601 GKDAGRFSAAWQLYKAQEELIKVAKEYGVKLTMFHGRGGTVGRGGGPTHLAILSQPPDTI 660
GKDAGRFSAAWQLYKAQEELI VAK++GVKLTMFHGRGGTVGRGGGPTHLAILSQPPDTI
Sbjct: 1843 GKDAGRFSAAWQLYKAQEELINVAKKFGVKLTMFHGRGGTVGRGGGPTHLAILSQPPDTI 2022
Query: 661 HGSLRVTVQGEVIEQSFGEEHLCFRTLQRYTAATLEHGMHPPISPKPEWRALMDEMAVIA 720
HGSLRVTVQGEVIEQSFGE+HLCFRTLQR+TAATLEHGMHPPISPKPEWRALMD+MAVIA
Sbjct: 2023 HGSLRVTVQGEVIEQSFGEQHLCFRTLQRFTAATLEHGMHPPISPKPEWRALMDQMAVIA 2202
Query: 721 TEEYRSIVFKEPRFVEYFRLATPELEYGRMNIGSRPAKRKPSGGIETLRAIPWIFAWTQT 780
TEEYRSIVFKEPRFVEYFRLATPELEYGRMNIGSRPAKR+PSGGIETLRAIPWIFAWTQT
Sbjct: 2203 TEEYRSIVFKEPRFVEYFRLATPELEYGRMNIGSRPAKRRPSGGIETLRAIPWIFAWTQT 2382
Query: 781 RFHLPVWLGFGAAFKHVIAKDIRNLNMLQEMYNQWPFFRVTIDLVEMVFAKGDPGIAALY 840
RFHLPVWLGFGAAFK VI ++++NLNMLQEMYNQWPFFRVT+DLVEMVFAKGDP IAAL
Sbjct: 2383 RFHLPVWLGFGAAFKKVIEENVKNLNMLQEMYNQWPFFRVTLDLVEMVFAKGDPKIAALN 2562
Query: 841 DRLLVSEDLWSFGEQLRTMFEETKQLLLQVAAHKDLLEGDPYLKQRLRLRDSYITTLNVC 900
DRLLVS+DLW FG+QLR +EET++LLLQVA HK++LEGDPYLKQRLRLR + ITTLN+
Sbjct: 2563 DRLLVSKDLWPFGDQLRNKYEETRKLLLQVAGHKEILEGDPYLKQRLRLRHAPITTLNIV 2742
Query: 901 QAYTLKRIRDPNYNVKLRPHISKEAIDVSKPADELVTLNPTSEYAPGLEDTLILTMKGIA 960
QAYTLKRIRDPNYNVK+RP ISKE+ + SK ADELV LNPTSEYAPGLEDTLILTMKGIA
Sbjct: 2743 QAYTLKRIRDPNYNVKVRPRISKESAEASKSADELVKLNPTSEYAPGLEDTLILTMKGIA 2922
Query: 961 AGMQNTG 967
AGMQNTG
Sbjct: 2923 AGMQNTG 2943
>TC206170 UP|CAP1_SOYBN (Q02909) Phosphoenolpyruvate carboxylase,
housekeeping isozyme (PEPCase) , complete
Length = 3286
Score = 1754 bits (4544), Expect = 0.0
Identities = 884/981 (90%), Positives = 918/981 (93%), Gaps = 14/981 (1%)
Frame = +1
Query: 1 MANRNLEKMASIDAQLRQLVPAKVSEDDKLVEYDALLLDRFLDILQDLHGEDLKETVQEV 60
MANRNLEKMASIDAQLR LVPAKVSEDDKLVEYDALLLDRFLDILQDLHGEDLKETVQEV
Sbjct: 1 MANRNLEKMASIDAQLRLLVPAKVSEDDKLVEYDALLLDRFLDILQDLHGEDLKETVQEV 180
Query: 61 YELSAEYERKHEPQRLEVLGNLITSLDAGDSIVVAKSFAHMLNLANLAEEVQIAHRRRIK 120
YELSAEYE KH+P++LE LGNLITSLDAGDSIVVAKSF+HMLNLANLAEEVQIAH RR K
Sbjct: 181 YELSAEYEGKHDPKKLEELGNLITSLDAGDSIVVAKSFSHMLNLANLAEEVQIAHSRRNK 360
Query: 121 LKKGDFADEGNATTESDIEETFKKLVGEMKKSPQEVFDELKNQTVDLVLTAHPTQSVRRS 180
LKKGDFADE NATTESDIEET KKLV +MKKSPQEVFD LKNQTVDLVLTAHPTQSVRRS
Sbjct: 361 LKKGDFADENNATTESDIEETLKKLVVDMKKSPQEVFDALKNQTVDLVLTAHPTQSVRRS 540
Query: 181 LLQKHGRIRNCLTQLYAKDITPDDKQELDEALQREIQAAFRTDEIRRTAPTPQDEMRAGM 240
LLQKHGRIRN LTQLYAKDITPDDKQELDEALQREIQAAFRTDEIRRT PTPQDEMRAGM
Sbjct: 541 LLQKHGRIRNNLTQLYAKDITPDDKQELDEALQREIQAAFRTDEIRRTPPTPQDEMRAGM 720
Query: 241 SYFHETIWKGVPKFLRRVDTALKNIGINERVPYNAPLIQFSSWMGGDRDGNPRVTPEVTR 300
SYFHETIWKGVP FLRRVDTALKNIGINERVPYNAPLIQFSSWMGGDRDGNPRVTPEVTR
Sbjct: 721 SYFHETIWKGVPTFLRRVDTALKNIGINERVPYNAPLIQFSSWMGGDRDGNPRVTPEVTR 900
Query: 301 DVCLLARMMAANLYYSQIEDLMFELSMWRCNDELRVRA-------EELTSSSNKDAVAK- 352
DVCLLARMMAANLYYSQIEDLMFELSMWRCNDELRV +E S + +
Sbjct: 901 DVCLLARMMAANLYYSQIEDLMFELSMWRCNDELRVPCR*T*QVFQEKFSRKTLHRILES 1080
Query: 353 ------HYIEFWKNIPPSEPYRVVLGEVRNRLYQTRERSRHLLANEYSDILEEHTFTNVE 406
H + +W ++RNRLYQTRERSRHLLA+ YSDI EE TFTNVE
Sbjct: 1081PFLQMNHIVCYWV-------------KLRNRLYQTRERSRHLLAHGYSDIPEEETFTNVE 1221
Query: 407 EFLEPLELCYRSLCACGDRAIADGSLLDFLRQVSTFGLSLVRLDIRQESDRHTDVLDAIT 466
EFLEPLELCYRSLCACGDRAIADGSLLDFLRQVSTFGLSLVRLDIRQESDRHTDVLDAIT
Sbjct: 1222EFLEPLELCYRSLCACGDRAIADGSLLDFLRQVSTFGLSLVRLDIRQESDRHTDVLDAIT 1401
Query: 467 KHLGIGSYLEWSEEKRQQWLLSELSGKRPLFGPDLPQTEEIKDVLDTFHVIAELPSDNFG 526
KHL IGSY EWSEEKRQQWLLSELSGKRPLFGPDLPQTEEI+DVL+TFHVIAELP DNFG
Sbjct: 1402KHLEIGSYQEWSEEKRQQWLLSELSGKRPLFGPDLPQTEEIRDVLETFHVIAELPLDNFG 1581
Query: 527 AYIISMATAPSDVLAVELLQRECHVKNPLRVVPLFEKLADLETAPAALARLFSVEWYRNR 586
AYIISMATAPSDVLAVELLQRECHVK+PLRVVPLFEKLADLE APAALARLFSV+WYRNR
Sbjct: 1582AYIISMATAPSDVLAVELLQRECHVKHPLRVVPLFEKLADLEAAPAALARLFSVDWYRNR 1761
Query: 587 INGKQEVMIGYSDSGKDAGRFSAAWQLYKAQEELIKVAKEYGVKLTMFHGRGGTVGRGGG 646
INGKQEVMIGYSDSGKDAGRFSAAWQLYKAQEELI VAK+YGVKLTMFHGRGGTVGRGGG
Sbjct: 1762INGKQEVMIGYSDSGKDAGRFSAAWQLYKAQEELIMVAKQYGVKLTMFHGRGGTVGRGGG 1941
Query: 647 PTHLAILSQPPDTIHGSLRVTVQGEVIEQSFGEEHLCFRTLQRYTAATLEHGMHPPISPK 706
PTHLAILSQPP+TIHGSLRVTVQGEVIEQSFGE+HLCFRTLQR+TAATLEHGMHPPISPK
Sbjct: 1942PTHLAILSQPPETIHGSLRVTVQGEVIEQSFGEQHLCFRTLQRFTAATLEHGMHPPISPK 2121
Query: 707 PEWRALMDEMAVIATEEYRSIVFKEPRFVEYFRLATPELEYGRMNIGSRPAKRKPSGGIE 766
PEWRALMDEMAVIATEEYRSIVFKEPRFVEYFRLATPELEYGRMNIGSRPAKR+PSGGIE
Sbjct: 2122PEWRALMDEMAVIATEEYRSIVFKEPRFVEYFRLATPELEYGRMNIGSRPAKRRPSGGIE 2301
Query: 767 TLRAIPWIFAWTQTRFHLPVWLGFGAAFKHVIAKDIRNLNMLQEMYNQWPFFRVTIDLVE 826
TLRAIPWIFAWTQTRFHLPVWLGFGAAFKHVI KD+RN+++LQEMYNQWPFFRVTIDLVE
Sbjct: 2302TLRAIPWIFAWTQTRFHLPVWLGFGAAFKHVIEKDVRNIHVLQEMYNQWPFFRVTIDLVE 2481
Query: 827 MVFAKGDPGIAALYDRLLVSEDLWSFGEQLRTMFEETKQLLLQVAAHKDLLEGDPYLKQR 886
MVFAKGDPGIAALYDRLLVSEDLWSFGEQLRTM+EETK+LLLQVA H+DLLEGDPYLKQR
Sbjct: 2482MVFAKGDPGIAALYDRLLVSEDLWSFGEQLRTMYEETKELLLQVAGHRDLLEGDPYLKQR 2661
Query: 887 LRLRDSYITTLNVCQAYTLKRIRDPNYNVKLRPHISKEAIDVSKPADELVTLNPTSEYAP 946
LRLRDSYITTLNVCQAYTLKRIRDPNYNVKLRPHISKE+I++SKPADEL+TLNPTSEYAP
Sbjct: 2662LRLRDSYITTLNVCQAYTLKRIRDPNYNVKLRPHISKESIEISKPADELITLNPTSEYAP 2841
Query: 947 GLEDTLILTMKGIAAGMQNTG 967
GLEDTLILTMKGIAAG+QNTG
Sbjct: 2842GLEDTLILTMKGIAAGLQNTG 2904
>TC217081 homologue to UP|Q6Q2Z8 (Q6Q2Z8) Phosphoenolpyruvate carboxylase ,
partial (42%)
Length = 1423
Score = 756 bits (1953), Expect = 0.0
Identities = 365/407 (89%), Positives = 392/407 (95%)
Frame = +3
Query: 561 FEKLADLETAPAALARLFSVEWYRNRINGKQEVMIGYSDSGKDAGRFSAAWQLYKAQEEL 620
FEKLADLE APAA+ARLFS++WYR+RINGKQEVMIGYSDSGKDAGRFSAAW LYKAQEEL
Sbjct: 3 FEKLADLEAAPAAVARLFSIDWYRDRINGKQEVMIGYSDSGKDAGRFSAAWALYKAQEEL 182
Query: 621 IKVAKEYGVKLTMFHGRGGTVGRGGGPTHLAILSQPPDTIHGSLRVTVQGEVIEQSFGEE 680
IKVAKE+GVKLTMFHGRGGTVGRGGGPTHLAILSQPPDTIHGSLRVTVQGEVIEQSFGEE
Sbjct: 183 IKVAKEFGVKLTMFHGRGGTVGRGGGPTHLAILSQPPDTIHGSLRVTVQGEVIEQSFGEE 362
Query: 681 HLCFRTLQRYTAATLEHGMHPPISPKPEWRALMDEMAVIATEEYRSIVFKEPRFVEYFRL 740
HLCFRTLQR+TAATLEHGMHPP++PKPEWRALMDEMAVIATEEYRSIVF+EPRFVEYFR
Sbjct: 363 HLCFRTLQRFTAATLEHGMHPPVAPKPEWRALMDEMAVIATEEYRSIVFQEPRFVEYFRC 542
Query: 741 ATPELEYGRMNIGSRPAKRKPSGGIETLRAIPWIFAWTQTRFHLPVWLGFGAAFKHVIAK 800
ATPELEYGRMNIGSRP+KRKPSGGIE+LRAIPWIFAWTQTRFHLPVWLGFGAAF HVI K
Sbjct: 543 ATPELEYGRMNIGSRPSKRKPSGGIESLRAIPWIFAWTQTRFHLPVWLGFGAAFSHVIKK 722
Query: 801 DIRNLNMLQEMYNQWPFFRVTIDLVEMVFAKGDPGIAALYDRLLVSEDLWSFGEQLRTMF 860
D +NL MLQ+MYNQWPFFRV++DLVEMVFAKGDPGIAALYD+LLVSE+LW FGE+LR+MF
Sbjct: 723 DPKNLQMLQDMYNQWPFFRVSLDLVEMVFAKGDPGIAALYDKLLVSEELWPFGERLRSMF 902
Query: 861 EETKQLLLQVAAHKDLLEGDPYLKQRLRLRDSYITTLNVCQAYTLKRIRDPNYNVKLRPH 920
EETK LLLQVA HKDLLEGDPYLKQRLRLRDSYITTLNV QAYTLKRIRDP+Y+VKLRPH
Sbjct: 903 EETKSLLLQVAGHKDLLEGDPYLKQRLRLRDSYITTLNVLQAYTLKRIRDPDYHVKLRPH 1082
Query: 921 ISKEAIDVSKPADELVTLNPTSEYAPGLEDTLILTMKGIAAGMQNTG 967
+SK+ ++ +KPA ELV LNPTS+YAPGLEDTLILTMKGIAAGMQNTG
Sbjct: 1083LSKDYMESNKPAAELVKLNPTSDYAPGLEDTLILTMKGIAAGMQNTG 1223
>TC208583 Phosphoenolpyruvate carboxylase [Glycine max]
Length = 1339
Score = 702 bits (1812), Expect = 0.0
Identities = 339/383 (88%), Positives = 363/383 (94%)
Frame = +2
Query: 585 NRINGKQEVMIGYSDSGKDAGRFSAAWQLYKAQEELIKVAKEYGVKLTMFHGRGGTVGRG 644
NRI+GKQEVMIGYSDSGKDAGR SAAW LYKAQEEL+KVAKEYGVKLTMFHGRGGTVGRG
Sbjct: 2 NRIDGKQEVMIGYSDSGKDAGRLSAAWALYKAQEELVKVAKEYGVKLTMFHGRGGTVGRG 181
Query: 645 GGPTHLAILSQPPDTIHGSLRVTVQGEVIEQSFGEEHLCFRTLQRYTAATLEHGMHPPIS 704
GGPTHLAILSQPPDTIHGSLRVTVQGEVIEQSFGEEHLCFRTLQR+TAATLEHGMHPP+S
Sbjct: 182 GGPTHLAILSQPPDTIHGSLRVTVQGEVIEQSFGEEHLCFRTLQRFTAATLEHGMHPPVS 361
Query: 705 PKPEWRALMDEMAVIATEEYRSIVFKEPRFVEYFRLATPELEYGRMNIGSRPAKRKPSGG 764
PKPEWRALMDEMAVIAT+EYRS+VFKEPRFVEYFR ATPELEYGRMNIGSRP+KRKPSGG
Sbjct: 362 PKPEWRALMDEMAVIATKEYRSVVFKEPRFVEYFRCATPELEYGRMNIGSRPSKRKPSGG 541
Query: 765 IETLRAIPWIFAWTQTRFHLPVWLGFGAAFKHVIAKDIRNLNMLQEMYNQWPFFRVTIDL 824
IE+LRAIPWIFAWTQTRFHLPVWLGFG+AFKHV+ KD +NL MLQ+MYNQWPFFRVT+DL
Sbjct: 542 IESLRAIPWIFAWTQTRFHLPVWLGFGSAFKHVVEKDPKNLQMLQDMYNQWPFFRVTLDL 721
Query: 825 VEMVFAKGDPGIAALYDRLLVSEDLWSFGEQLRTMFEETKQLLLQVAAHKDLLEGDPYLK 884
VEMVFAKGDPGIAAL+D+LLVSE+L FGE LR +EETK LLQVA HKD+LEGDPYLK
Sbjct: 722 VEMVFAKGDPGIAALFDKLLVSEELRPFGENLRAKYEETKSFLLQVAGHKDILEGDPYLK 901
Query: 885 QRLRLRDSYITTLNVCQAYTLKRIRDPNYNVKLRPHISKEAIDVSKPADELVTLNPTSEY 944
QRLRLRDSYITTLNV QAYTLKRIRDP+Y+VKLRPH+SK+ ++ SKPA ELV LNP SEY
Sbjct: 902 QRLRLRDSYITTLNVLQAYTLKRIRDPDYHVKLRPHLSKDYMESSKPAAELVKLNPKSEY 1081
Query: 945 APGLEDTLILTMKGIAAGMQNTG 967
APGLEDTLILTMKGIAAGMQNTG
Sbjct: 1082APGLEDTLILTMKGIAAGMQNTG 1150
>TC227184 UP|Q6Q2Z9 (Q6Q2Z9) Phosphoenolpyruvate carboxylase , partial (63%)
Length = 2132
Score = 544 bits (1402), Expect = e-155
Identities = 288/641 (44%), Positives = 400/641 (61%), Gaps = 16/641 (2%)
Frame = +1
Query: 343 SSSNKDAVAKHYIEFWKNIPPSEPYRVVLGEVRNRLYQTRERSRHLLANEYSDILEEHTF 402
S + K K +P PYR+VLG V+++L ++R R LL + D +
Sbjct: 121 SQTGKSTFQKLLEPMLPQLPGIAPYRIVLGNVKDKLEKSRRRLEILLEDVACDYDPLDYY 300
Query: 403 TNVEEFLEPLELCYRSLCACGDRAIADGSLLDFLRQVSTFGLSLVRLDIRQESDRHTDVL 462
++ LEPL LCY SL +CG +ADG L D +R+V+TFG+ L++LD+RQES RH + L
Sbjct: 301 ETSDQLLEPLLLCYESLQSCGSGVLADGRLADLIRRVATFGMVLMKLDLRQESGRHAEAL 480
Query: 463 DAITKHLGIGSYLEWSEEKRQQWLLSELSGKRPLFGPDLPQTEEIKDVLDTFHVIAELPS 522
DAIT++L +G+Y EW EEK+ +L EL GKRPL + ++K+VLDTF + AEL S
Sbjct: 481 DAITQYLDMGTYSEWDEEKKLDFLTRELKGKRPLVPVSIEVHPDVKEVLDTFRIAAELGS 660
Query: 523 DNFGAYIISMATAPSDVLAVELLQRECHVK-----------NPLRVVPLFEKLADLETAP 571
D+ GAY+ISMA+ SDVLAVELLQ++ + LRVVPLFE + DL A
Sbjct: 661 DSLGAYVISMASNASDVLAVELLQKDARLAAIGELGKACPGGTLRVVPLFETVKDLRGAG 840
Query: 572 AALARLFSVEWYRNRI----NGKQEVMIGYSDSGKDAGRFSAAWQLYKAQEELIKVAKEY 627
+ + +L S++WY I NG QEVM+GYSDSGKDAGRF+AAW+LYKAQE+++ +Y
Sbjct: 841 SVIRKLLSIDWYHEHIIKNHNGHQEVMVGYSDSGKDAGRFTAAWELYKAQEDVVAACNDY 1020
Query: 628 GVKLTMFHGRGGTVGRGGGPTHLAILSQPPDTIHGSLRVTVQGEVIEQSFGEEHLCFRTL 687
G+K+T+FHGRGG++GRGGGPT+LAI SQPP ++ G+LR T QGE++E FG + R L
Sbjct: 1021GIKVTLFHGRGGSIGRGGGPTYLAIQSQPPGSVMGTLRSTEQGEMVEAKFGLPQIAVRQL 1200
Query: 688 QRYTAATLEHGMHPPISPKPE-WRALMDEMAVIATEEYRSIVFKEPRFVEYFRLATPELE 746
+ YT A L + PPI P+ E WR +M+E++ I+ + YR++V++ P F+ YF ATPE E
Sbjct: 1201EIYTTAVLLATLRPPIPPREEKWRNVMEEISNISCQCYRNVVYENPEFLAYFHEATPEAE 1380
Query: 747 YGRMNIGSRPAKRKPSGGIETLRAIPWIFAWTQTRFHLPVWLGFGAAFKHVIAKDIRNLN 806
G +NIGSRP +RK S GI LRAIPW+FAWTQTRF LP WLG GA K K
Sbjct: 1381LGFLNIGSRPTRRKSSVGIGHLRAIPWLFAWTQTRFVLPAWLGVGAGLKGACEKGY--TE 1554
Query: 807 MLQEMYNQWPFFRVTIDLVEMVFAKGDPGIAALYDRLLVSEDLWSFGEQLRTMFEETKQL 866
L+ MY +WPFF+ TIDL+EMV K D IA YD +LVS++ G +LR+ ++
Sbjct: 1555ELKAMYKEWPFFQSTIDLIEMVLGKADIPIAKHYDEVLVSKERQELGHELRSELMTAEKF 1734
Query: 867 LLQVAAHKDLLEGDPYLKQRLRLRDSYITTLNVCQAYTLKRIRDPNYNVKLRPHISKEAI 926
++ ++ H+ L + + L++ + R ++ LN+ Q LKR+R + N K+R
Sbjct: 1735VMVISGHEKLQQNNRSLRRLIENRLPFLNPLNMLQVEILKRLRRDDDNRKIR-------- 1890
Query: 927 DVSKPADELVTLNPTSEYAPGLEDTLILTMKGIAAGMQNTG 967
D L++T+ GIAAGM+NTG
Sbjct: 1891-----------------------DALLITINGIAAGMKNTG 1944
>TC220940 homologue to UP|Q9T0N1 (Q9T0N1) Ppc2 protein , partial (16%)
Length = 480
Score = 275 bits (704), Expect = 5e-74
Identities = 134/159 (84%), Positives = 148/159 (92%)
Frame = +3
Query: 366 PYRVVLGEVRNRLYQTRERSRHLLANEYSDILEEHTFTNVEEFLEPLELCYRSLCACGDR 425
PYRV+LG+VR++LY TRER+R LLAN S+I EE TFTNVE+FLEPLELCYRSLCACGD+
Sbjct: 3 PYRVILGDVRDKLYNTRERARQLLANGSSEIPEETTFTNVEQFLEPLELCYRSLCACGDQ 182
Query: 426 AIADGSLLDFLRQVSTFGLSLVRLDIRQESDRHTDVLDAITKHLGIGSYLEWSEEKRQQW 485
IADGSLLDFLRQVSTFGLSLVRLDIRQESDRHTDV+DAIT HL IGSY EWSEE+RQ+W
Sbjct: 183 PIADGSLLDFLRQVSTFGLSLVRLDIRQESDRHTDVMDAITNHLEIGSYREWSEERRQEW 362
Query: 486 LLSELSGKRPLFGPDLPQTEEIKDVLDTFHVIAELPSDN 524
LLSELSGKRPLFGPDLP+TEEI DVL+TFHVIAELPSD+
Sbjct: 363 LLSELSGKRPLFGPDLPKTEEIADVLETFHVIAELPSDS 479
>CA783206 similar to PIR|T09846|T09 phosphoenolpyruvate carboxylase (EC
4.1.1.31) 1 - upland cotton, partial (14%)
Length = 450
Score = 244 bits (622), Expect = 2e-64
Identities = 123/141 (87%), Positives = 135/141 (95%), Gaps = 1/141 (0%)
Frame = +2
Query: 79 LGNLITSLDAGDSIVVAKSFAHMLNLANLAEEVQIAHRRRIKL-KKGDFADEGNATTESD 137
LGN++T LDAGDSIV++KSFAHMLNLANLAEEVQIA+RRRIKL KKGDFADE +A TESD
Sbjct: 26 LGNMLTGLDAGDSIVISKSFAHMLNLANLAEEVQIAYRRRIKLLKKGDFADENSAITESD 205
Query: 138 IEETFKKLVGEMKKSPQEVFDELKNQTVDLVLTAHPTQSVRRSLLQKHGRIRNCLTQLYA 197
IEETFK+LV ++KK+PQE+FD LK+QTVDLVLTAHPTQSVRRSLLQKHGRIRNCLTQLYA
Sbjct: 206 IEETFKRLVNQLKKTPQEIFDALKSQTVDLVLTAHPTQSVRRSLLQKHGRIRNCLTQLYA 385
Query: 198 KDITPDDKQELDEALQREIQA 218
KDITPDDKQELDEALQREIQA
Sbjct: 386 KDITPDDKQELDEALQREIQA 448
>BU577401 homologue to EGAD|139142|14 ppc2 protein {Mesembryanthemum
crystallinum}, partial (13%)
Length = 366
Score = 226 bits (577), Expect = 3e-59
Identities = 107/123 (86%), Positives = 114/123 (91%)
Frame = +1
Query: 239 GMSYFHETIWKGVPKFLRRVDTALKNIGINERVPYNAPLIQFSSWMGGDRDGNPRVTPEV 298
GMSYFHETIWKG+PKFLRRVDTALKNIGINERVPYNAP+IQFSSWMGGDRDGNPRVTPEV
Sbjct: 4 GMSYFHETIWKGIPKFLRRVDTALKNIGINERVPYNAPVIQFSSWMGGDRDGNPRVTPEV 183
Query: 299 TRDVCLLARMMAANLYYSQIEDLMFELSMWRCNDELRVRAEELTSSSNKDAVAKHYIEFW 358
TRDVC +RMMAANLY+SQIEDLMFELSMW CNDELR R++EL SSS +D AKHYIEFW
Sbjct: 184 TRDVCFXSRMMAANLYFSQIEDLMFELSMWSCNDELRDRSDELLSSSKRD--AKHYIEFW 357
Query: 359 KNI 361
K I
Sbjct: 358 KQI 366
>AW704161 weakly similar to GP|18073660|em phosphoenolpyruvate carboxylase
{Sticherus bifidus}, partial (43%)
Length = 595
Score = 221 bits (563), Expect = 1e-57
Identities = 109/198 (55%), Positives = 143/198 (72%)
Frame = +2
Query: 631 LTMFHGRGGTVGRGGGPTHLAILSQPPDTIHGSLRVTVQGEVIEQSFGEEHLCFRTLQRY 690
LTMFHGRGGTVGRG PTHLAILSQPPDTIH SL +T+QG+VI+ SF E+HL F+TLQ +
Sbjct: 2 LTMFHGRGGTVGRGRSPTHLAILSQPPDTIHSSLAITIQGKVIKHSFKEKHLYFKTLQHF 181
Query: 691 TAATLEHGMHPPISPKPEWRALMDEMAVIATEEYRSIVFKEPRFVEYFRLATPELEYGRM 750
T TL+H +HPPI+PKP+ R L++ + +I T+ Y SI+F++P F+EYF+ TP+L Y +
Sbjct: 182 TTTTLKHKIHPPITPKPQ*RTLINNITIITTQNYHSIIFQKPHFIEYFQYTTPKLYYRII 361
Query: 751 NIGSRPAKRKPSGGIETLRAIPWIFAWTQTRFHLPVWLGFGAAFKHVIAKDIRNLNMLQE 810
NI S P+K KPS I++L+AIP I TQT HLP+ L A H+I K+ R+L LQ
Sbjct: 362 NINSHPSKLKPSRKIKSLQAIP*ICT*TQTEVHLPI*LSL*TAISHIINKNPRDLQRLQH 541
Query: 811 MYNQWPFFRVTIDLVEMV 828
MY+Q ++ L EMV
Sbjct: 542 MYDQ*RLRQIRRYLGEMV 595
>NP004804 phosphoenolpyruvate carboxylase
Length = 180
Score = 95.9 bits (237), Expect = 8e-20
Identities = 49/60 (81%), Positives = 55/60 (91%), Gaps = 1/60 (1%)
Frame = +1
Query: 95 AKSFAHMLNLANLAEEVQIAHRRRIKL-KKGDFADEGNATTESDIEETFKKLVGEMKKSP 153
AKSF+HMLNLANLAEEVQIA+RRRIKL KKGDF DE +A TESDIEETFKKLV ++KK+P
Sbjct: 1 AKSFSHMLNLANLAEEVQIAYRRRIKLLKKGDFPDENSAITESDIEETFKKLVAQLKKTP 180
>TC210689 homologue to UP|Q6Q2Z9 (Q6Q2Z9) Phosphoenolpyruvate carboxylase ,
partial (7%)
Length = 948
Score = 91.7 bits (226), Expect = 1e-18
Identities = 40/75 (53%), Positives = 57/75 (75%)
Frame = +3
Query: 427 IADGSLLDFLRQVSTFGLSLVRLDIRQESDRHTDVLDAITKHLGIGSYLEWSEEKRQQWL 486
+ADG L D +R+V+TFG+ L++LD+RQES RH + +DAIT++L +G+Y EW EEK+ +L
Sbjct: 6 LADGRLADLIRRVATFGMVLMKLDLRQESSRHAETIDAITRYLDMGTYSEWDEEKKLDFL 185
Query: 487 LSELSGKRPLFGPDL 501
EL GKRPL P +
Sbjct: 186 TRELKGKRPLVPPSI 230
>BM309173
Length = 374
Score = 89.7 bits (221), Expect = 5e-18
Identities = 39/73 (53%), Positives = 55/73 (74%)
Frame = +1
Query: 429 DGSLLDFLRQVSTFGLSLVRLDIRQESDRHTDVLDAITKHLGIGSYLEWSEEKRQQWLLS 488
DG L D +R+V+TFG+ L++LD+RQES RH + +DAIT++L +G+Y EW EEK+ +L
Sbjct: 1 DGQLADLIRRVATFGMVLMKLDLRQESSRHAETIDAITRYLDMGTYSEWDEEKKLDFLTR 180
Query: 489 ELSGKRPLFGPDL 501
EL GKRPL P +
Sbjct: 181 ELKGKRPLVPPSI 219
>BM086497
Length = 428
Score = 81.6 bits (200), Expect = 1e-15
Identities = 36/70 (51%), Positives = 52/70 (73%)
Frame = +3
Query: 437 RQVSTFGLSLVRLDIRQESDRHTDVLDAITKHLGIGSYLEWSEEKRQQWLLSELSGKRPL 496
R+V+TFG+ L++LD+RQES RH + +DAIT++L +G+Y EW EEK+ +L EL GKRPL
Sbjct: 3 RRVATFGMVLMKLDLRQESSRHAETIDAITRYLDMGTYSEWDEEKKLDFLTRELKGKRPL 182
Query: 497 FGPDLPQTEE 506
P + + E
Sbjct: 183 VPPSIEERFE 212
>TC208584 homologue to UP|O22119 (O22119) Phosphoenolpyruvate carboxylase
(Fragment) , complete
Length = 317
Score = 71.2 bits (173), Expect = 2e-12
Identities = 35/47 (74%), Positives = 40/47 (84%)
Frame = +2
Query: 921 ISKEAIDVSKPADELVTLNPTSEYAPGLEDTLILTMKGIAAGMQNTG 967
+SK+ ++ SKPA ELV +NP SEYAPGLE TLILTMKGIAAGM NTG
Sbjct: 2 LSKDYMESSKPAAELVKVNPKSEYAPGLEXTLILTMKGIAAGMXNTG 142
>AW705942 weakly similar to GP|25901015|dbj phosphoenolpyruvate carboxylase
{Glycine max}, partial (6%)
Length = 181
Score = 68.6 bits (166), Expect = 1e-11
Identities = 34/60 (56%), Positives = 41/60 (67%)
Frame = +2
Query: 784 LPVWLGFGAAFKHVIAKDIRNLNMLQEMYNQWPFFRVTIDLVEMVFAKGDPGIAALYDRL 843
LPVWLGFGA+FK I +++ NL + E QWP R T+ LV MVFA GD +AAL DRL
Sbjct: 2 LPVWLGFGASFKKGIDENVNNLCLRNETCKQWPCCRATLHLVAMVFAGGDSKLAALNDRL 181
>BI468874
Length = 422
Score = 55.8 bits (133), Expect = 9e-08
Identities = 43/146 (29%), Positives = 77/146 (52%), Gaps = 1/146 (0%)
Frame = +2
Query: 79 LGNLITSLDAGDSIVVAKSFAHMLNLANLAEEVQIAHRRRIKLKKGDFADEGNATTES-D 137
L + ++ + +++ +A++F+H L L +AE H R ++KG GN +
Sbjct: 23 LASELSKMTLEEALPLARAFSHHLTLMGIAE----THHR---VRKG-----GNMVLAAKS 166
Query: 138 IEETFKKLVGEMKKSPQEVFDELKNQTVDLVLTAHPTQSVRRSLLQKHGRIRNCLTQLYA 197
++ F L+ + SP E++ + Q V++VLTAHPTQ R+L KH +I + L
Sbjct: 167 CDDIFNNLLQD-GVSPVELYKTVFKQEVEIVLTAHPTQINHRTLQYKHLKIAHLLDYND* 343
Query: 198 KDITPDDKQELDEALQREIQAAFRTD 223
D++P+D+ E L E + ++TD
Sbjct: 344 PDLSPEDRDMWIEDLVGEKTSIWQTD 421
>BF068053
Length = 202
Score = 49.7 bits (117), Expect = 6e-06
Identities = 27/66 (40%), Positives = 41/66 (61%), Gaps = 4/66 (6%)
Frame = +2
Query: 572 AALARLFSVEWYRNRI----NGKQEVMIGYSDSGKDAGRFSAAWQLYKAQEELIKVAKEY 627
+ + RL ++ Y I + Q+VM GYSD GKDAGR++A +LYKAQE++ +Y
Sbjct: 5 SCIRRLSLIDCYLEHIVENHHVHQKVMGGYSDFGKDAGRYTADRELYKAQEDVAACCLDY 184
Query: 628 GVKLTM 633
G +T+
Sbjct: 185 GHTVTL 202
>TC210666
Length = 796
Score = 31.6 bits (70), Expect = 1.8
Identities = 18/55 (32%), Positives = 23/55 (41%), Gaps = 2/55 (3%)
Frame = +2
Query: 758 KRKPSGGIETLRAIPWIFAWTQTRFHLPVWLGFGAAFKHVI--AKDIRNLNMLQE 810
K P G+ + W F W + HLP+W GFG H IRN+ E
Sbjct: 296 KPTPHEGVNV--GLQWKFEWN--KIHLPLWKGFGLHISHTYHGRAVIRNIETFME 448
>TC209778 similar to UP|Q88GI3 (Q88GI3) Major facilitator family transporter,
partial (4%)
Length = 837
Score = 27.3 bits (59), Expect(2) = 2.2
Identities = 11/38 (28%), Positives = 20/38 (51%)
Frame = +2
Query: 832 GDPGIAALYDRLLVSEDLWSFGEQLRTMFEETKQLLLQ 869
G ALY+ +L +D+W+ Q+ + TK+ L+
Sbjct: 560 GGDAFTALYNDVLWDDDIWNLNNQIPIPLDTTKRFHLE 673
Score = 22.3 bits (46), Expect(2) = 2.2
Identities = 10/26 (38%), Positives = 15/26 (57%)
Frame = +1
Query: 773 WIFAWTQTRFHLPVWLGFGAAFKHVI 798
W+FA T TR+H L G +++ I
Sbjct: 412 WVFAATSTRWHAD-GLELGGEYEYAI 486
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.319 0.136 0.399
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 38,330,124
Number of Sequences: 63676
Number of extensions: 503414
Number of successful extensions: 2324
Number of sequences better than 10.0: 46
Number of HSP's better than 10.0 without gapping: 2300
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2316
length of query: 967
length of database: 12,639,632
effective HSP length: 106
effective length of query: 861
effective length of database: 5,889,976
effective search space: 5071269336
effective search space used: 5071269336
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 64 (29.3 bits)
Lotus: description of TM0340.18


