

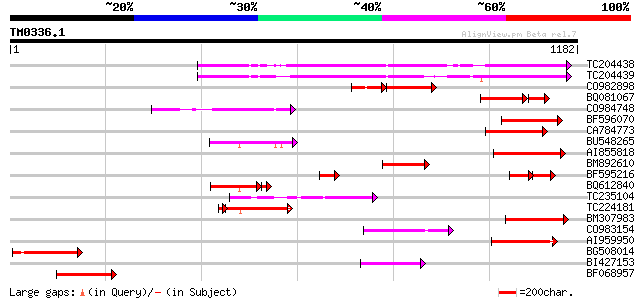
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0336.1
(1182 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC204438 homologue to UP|Q84VH6 (Q84VH6) Gag-pol polyprotein, co... 314 2e-85
TC204439 UP|Q84VI4 (Q84VI4) Gag-pol polyprotein, complete 314 2e-85
CO982898 117 2e-49
BQ081067 weakly similar to GP|23495377|dbj orf490 {Oryza sativa ... 131 4e-44
CO984748 174 2e-43
BF596070 similar to GP|27901698|gb gag-pol polyprotein {Vitis vi... 160 3e-39
CA784773 weakly similar to GP|27901698|gb gag-pol polyprotein {V... 157 2e-38
BU548265 homologue to GP|18000935|gb| cytochrome b {Mabuya delal... 153 5e-37
AI855818 weakly similar to GP|21741393|e OSJNBb0051N19.6 {Oryza ... 144 3e-34
BM892610 141 1e-33
BF595216 79 2e-31
BQ612840 120 3e-29
TC235104 weakly similar to UP|Q850H8 (Q850H8) Gag-pol polyprotei... 122 7e-28
TC224181 116 2e-27
BM307983 120 4e-27
CO983154 117 3e-26
AI959950 115 1e-25
BG508014 GP|15128578|db prolactin receptor {Paralichthys olivace... 112 1e-24
BI427153 111 2e-24
BF068957 weakly similar to PIR|B85188|B85 retrotransposon like p... 110 5e-24
>TC204438 homologue to UP|Q84VH6 (Q84VH6) Gag-pol polyprotein, complete
Length = 4734
Score = 314 bits (804), Expect = 2e-85
Identities = 243/797 (30%), Positives = 369/797 (45%), Gaps = 16/797 (2%)
Frame = +1
Query: 391 GVPYRGPAPSPMG----YAQAHVAFQTSTSTPQATSPAQSWYPDSGASHHVTSESQNVHQ 446
G P+ G S G + H T S + WY DSG S H+T + +
Sbjct: 1567 GHPHHGTQSSSSGRKMMWVPKHKIVSLVVHTSLRASAKEDWYLDSGCSRHMTGVKEFLVN 1746
Query: 447 FTPFAGQDQLFVGNGQGLPISGIGSSTLSSGLVLNNLLHVPSITKNLVSVSKFARDNGVF 506
P + F G+G I+G+G LN +L V +T NL+S+S+ D G
Sbjct: 1747 IEPCSTSYVTF-GDGSKGKITGMGKLVHDGLPSLNKVLLVKGLTANLISISQLC-DEGFN 1920
Query: 507 FEFHPYYCCVKSQASSKVLLQGSVGADGLYKFHEFKLSHLPVSLSAPVQHPVSVHTSVSS 566
F C V ++ S+VL++GS D Y L P + TS SS
Sbjct: 1921 VNFTKSECLVTNE-KSEVLMKGSRSKDNCY-------------LWTPQE------TSYSS 2040
Query: 567 SGHAPVEQPSVVSSVCTANLWHFRLGHPSLNVLRKALQFCKIPVSNKMDVD---FCKACV 623
+ + S +WH R GH L ++K + + + ++ C C
Sbjct: 2041 T--------CLFSKEDEVKIWHQRFGHLHLRGMKKIIDKGAVRGIPNLKIEEGRICGECQ 2196
Query: 624 MGKSHRLPSSLSTTEYTSP-LELVYSDLWGPSPSPSSSGYLYYITFIDAYSRYTWIYLLK 682
+GK ++ + TS LEL++ DL GP S G Y +D +SR+TW+ ++
Sbjct: 2197 IGKQVKMSHQKLQHQTTSRVLELLHMDLMGPMQVESLGGKRYAYVVVDDFSRFTWVNFIR 2376
Query: 683 AKSEAISAFHQFKASSELQLNAKIKALQTDWGGEFR--VFPNFLKDFGIHHRLICPHTHH 740
KS+ F + + + + IK +++D G EF F F GI H T
Sbjct: 2377 EKSDTFEVFKELSLRLQREKDCVIKRIRSDHGREFENSKFTEFCTSEGITHEFSAAITPQ 2556
Query: 741 QNGVVERKHRHIVDLGLSLLAHASIPLTYWDFAFSTAVYLINRLPTSSLQFDIP--LYKL 798
QNG+VERK+R + + +L +P W A +TA Y+ NR+ +L+ P LY++
Sbjct: 2557 QNGIVERKNRTLQEAARVMLHAKELPYNLWAEAMNTACYIHNRV---TLRRGTPTTLYEI 2727
Query: 799 FK-QIPDYTFLKVFGCTCFPLMRPYNSHKLDFRSQECVFLGYSPSHRGYKCLASSGRIYI 857
+K + P +FG C+ L K+D +S +FLGYS + R Y+ S R +
Sbjct: 2728 WKGRKPTVKHFHIFGSPCYILADREQRRKMDPKSDAGIFLGYSTNSRAYRVFNSRTRTVM 2907
Query: 858 -SKDVIFDEHTFPYPKLFAQNSNSPSLSPAVVLPSLPSVEHFSQPAIAQPSSSLPSPFLP 916
S +V+ D+ T K ++ + + A S + E+ S A +P+ + P
Sbjct: 2908 ESINVVVDDLTPARKKDVEEDVRTSGDNVADTAKSAENAEN-SDSATDEPNINQP----- 3069
Query: 917 SPIASSDQNPSSTSPASAPATSEASAEIPSQVTIPTSSGQVSAPSSSGNVHPMITRSKNG 976
D+ PS P E P++ + T S ++ S+S V
Sbjct: 3070 ------DKRPSIRIQKMHP--KELIIGDPNR-GVTTRSREIEIVSNSCFVS--------- 3195
Query: 977 IVQPRVQPTLLTVQLEPTTVKQALAHPQWLQAMQAEFDALQKNKTWSLVPLPPGREAIGC 1036
++EP VK+AL W+ AMQ E + ++N+ W LVP P G IG
Sbjct: 3196 -------------KIEPKNVKEALTDEFWINAMQEELEQFKRNEVWELVPRPEGTNVIGT 3336
Query: 1037 KWVFRVKHNPDGTLNKYKARLVAKGFHQQPGFDYTETFSPVIKPVTIRVILTLALTNHWT 1096
KW+F+ K N +G + + KARLVA+G+ Q G D+ ETF+PV + +IR++L +A +
Sbjct: 3337 KWIFKNKTNEEGVITRNKARLVAQGYTQIEGVDFDETFAPVARLESIRLLLGVACILKFK 3516
Query: 1097 IQQLDVNNAFLNGDLAEEVYMQQPQGF--EHDKKLVCRLHKALYGLKQAPRAWFEKLTSA 1154
+ Q+DV +AFLNG L EE Y++QP+GF V RL KALYGLKQAPRAW+E+LT
Sbjct: 3517 LYQMDVKSAFLNGYLNEEAYVEQPKGFVDPTHPDHVYRLKKALYGLKQAPRAWYERLTEF 3696
Query: 1155 LVQFGFVASKCDPSLFV 1171
L Q G+ D +LFV
Sbjct: 3697 LTQQGYRKGGIDKTLFV 3747
>TC204439 UP|Q84VI4 (Q84VI4) Gag-pol polyprotein, complete
Length = 4731
Score = 314 bits (804), Expect = 2e-85
Identities = 240/805 (29%), Positives = 360/805 (43%), Gaps = 24/805 (2%)
Frame = +1
Query: 391 GVPYRGPAPS----PMGYAQAHVAFQTSTSTPQATSPAQSWYPDSGASHHVTSESQNVHQ 446
G P+ G S M + H A T S + WY DSG S H+T + +
Sbjct: 1564 GHPHHGTQSSNSRKKMMWVPKHKAVSLVVHTSLRASAKEDWYLDSGCSRHMTGVKEFLLN 1743
Query: 447 FTPFAGQDQLFVGNGQGLPISGIGSSTLSSGLVLNNLLHVPSITKNLVSVSKFARDNGVF 506
P + F G+G I G+G LN +L V +T NL+S+S+ D G
Sbjct: 1744 IEPCSTSYVTF-GDGSKGKIIGMGKLVHDGLPSLNKVLLVKGLTANLISISQLC-DEGFN 1917
Query: 507 FEFHPYYCCVKSQASSKVLLQGSVGADGLYKFHEFKLSHLPVSLSAPVQHPVSVHTSVSS 566
F C V ++ S+VL++GS D Y + + S+ LS+
Sbjct: 1918 VNFTKSECLVTNE-KSEVLMKGSRSKDNCYLWTPQETSYSSTCLSSKEDE---------- 2064
Query: 567 SGHAPVEQPSVVSSVCTANLWHFRLGHPSLNVLRKALQFCKIPVSNKMDVD---FCKACV 623
+WH R GH L ++K + + + ++ C C
Sbjct: 2065 -----------------VRIWHQRFGHLHLRGMKKIIDKGAVRGIPNLKIEEGRICGECQ 2193
Query: 624 MGKSHRLPSSLSTTEYTSP-LELVYSDLWGPSPSPSSSGYLYYITFIDAYSRYTWIYLLK 682
+GK ++ + TS LEL++ DL GP S G Y +D +SR+TW+ ++
Sbjct: 2194 IGKQVKMSHQKLQHQTTSRVLELLHMDLMGPMQVESLGGKRYAYVVVDDFSRFTWVNFIR 2373
Query: 683 AKSEAISAFHQFKASSELQLNAKIKALQTDWGGEFRV--FPNFLKDFGIHHRLICPHTHH 740
KSE F + + + + IK +++D G EF F F GI H T
Sbjct: 2374 EKSETFEVFKELSLRLQREKDCVIKRIRSDHGREFENSRFTEFCTSEGITHEFSAAITPQ 2553
Query: 741 QNGVVERKHRHIVDLGLSLLAHASIPLTYWDFAFSTAVYLINRLPTSSLQFDIP--LYKL 798
QNG+VERK+R + + +L +P W A +TA Y+ NR+ +L+ P LY++
Sbjct: 2554 QNGIVERKNRTLQEAARVMLHAKELPYNLWAEAMNTACYIHNRV---TLRRGTPTTLYEI 2724
Query: 799 FK-QIPDYTFLKVFGCTCFPLMRPYNSHKLDFRSQECVFLGYSPSHRGYKCLASSGRIYI 857
+K + P +FG C+ L K+D +S +FLGYS + R Y+ S R +
Sbjct: 2725 WKGRKPSVKHFHIFGSPCYILADREQRRKMDPKSDAGIFLGYSTNSRAYRVFNSRTRTVM 2904
Query: 858 -SKDVIFDEHTFPYPKLFAQNSNSPSLSPAVVLPSLPSVEHFSQPAIAQPSSSLPSPFLP 916
S +V+ D+ LSPA +
Sbjct: 2905 ESINVVVDD-----------------LSPA------------------------RKKDVE 2961
Query: 917 SPIASSDQNPSSTSPASAPATSEASAEIPSQVTIPTSSGQVSAPSSSGNVHP--MITRSK 974
+ +S N + + + A + SA S + P +HP +I
Sbjct: 2962 EDVRTSGDNVADAAKSGENAENSDSATDESNINQPDKRSSTRIQK----MHPKELIIGDP 3129
Query: 975 NGIVQPR------VQPTLLTVQLEPTTVKQALAHPQWLQAMQAEFDALQKNKTWSLVPLP 1028
N V R V + ++EP VK+AL W+ AMQ E + ++N+ W LVP P
Sbjct: 3130 NRGVTTRSREVEIVSNSCFVSKIEPKNVKEALTDEFWINAMQEELEQFKRNEVWELVPRP 3309
Query: 1029 PGREAIGCKWVFRVKHNPDGTLNKYKARLVAKGFHQQPGFDYTETFSPVIKPVTIRVILT 1088
G IG KW+F+ K N +G + + KARLVA+G+ Q G D+ ETF+PV + +IR++L
Sbjct: 3310 EGTNVIGTKWIFKNKTNEEGVITRNKARLVAQGYTQIEGVDFDETFAPVARLESIRLLLG 3489
Query: 1089 LALTNHWTIQQLDVNNAFLNGDLAEEVYMQQPQGFEH--DKKLVCRLHKALYGLKQAPRA 1146
+A + + Q+DV +AFLNG L EEVY++QP+GF V RL KALYGLKQAPRA
Sbjct: 3490 VACILKFKLYQMDVKSAFLNGYLNEEVYVEQPKGFADPTHPDHVYRLKKALYGLKQAPRA 3669
Query: 1147 WFEKLTSALVQFGFVASKCDPSLFV 1171
W+E+LT L Q G+ D +LFV
Sbjct: 3670 WYERLTEFLTQQGYRKGGIDKTLFV 3744
>CO982898
Length = 816
Score = 117 bits (294), Expect(2) = 2e-49
Identities = 57/110 (51%), Positives = 77/110 (69%), Gaps = 4/110 (3%)
Frame = -1
Query: 785 PTSSLQFDIPLYKLFKQIPDYTFLKVFGCTCFPLMRPYNSHKLDFRSQECVFLGYSPSHR 844
PT S+ F +P +LF Q+ DY+F +VFGC CFPL+RPYN K +F+S EC+FLG SH+
Sbjct: 597 PTVSINFAVPYTQLFNQMLDYSFPRVFGCACFPLIRPYNIQKFNFKSHECIFLGNYTSHK 418
Query: 845 GYKCLASSGRIYISKDVIFDEHTFPYPKLFAQNSNS----PSLSPAVVLP 890
YKCLA +G++ +SKDVIF+E FPY +LF +S++ P PA V P
Sbjct: 417 NYKCLAPAGKL-VSKDVIFNEI*FPYKELFLPDSSTCIPVPQPIPATVPP 271
Score = 98.6 bits (244), Expect(2) = 2e-49
Identities = 49/74 (66%), Positives = 55/74 (74%)
Frame = -3
Query: 712 DWGGEFRVFPNFLKDFGIHHRLICPHTHHQNGVVERKHRHIVDLGLSLLAHASIPLTYWD 771
DWGGEF FL D GI H+LI PHT +QN VVERK RHIV GLSLL+HAS+PL +W
Sbjct: 814 DWGGEF*PLTEFLND*GIIHKLIFPHTQYQN-VVERKQRHIVKFGLSLLSHASLPLKFWG 638
Query: 772 FAFSTAVYLINRLP 785
AF T V+LINRLP
Sbjct: 637 HAFITPVFLINRLP 596
>BQ081067 weakly similar to GP|23495377|dbj orf490 {Oryza sativa (japonica
cultivar-group)}, partial (18%)
Length = 430
Score = 131 bits (330), Expect(2) = 4e-44
Identities = 63/98 (64%), Positives = 76/98 (77%)
Frame = +1
Query: 982 VQPTLLTVQLEPTTVKQALAHPQWLQAMQAEFDALQKNKTWSLVPLPPGREAIGCKWVFR 1041
+ PTLL EP+TVKQAL P W QAMQA+FDAL +NKT +L LP G+ AI CKWVFR
Sbjct: 1 LHPTLLFTTAEPSTVKQALISPPWRQAMQADFDALMENKTLTLTSLPSGKAAIDCKWVFR 180
Query: 1042 VKHNPDGTLNKYKARLVAKGFHQQPGFDYTETFSPVIK 1079
+K N GTLN+Y++RLVAKGFH + G DY+ETFSPVI+
Sbjct: 181 IKENLYGTLNRYRSRLVAKGFHLKFGCDYSETFSPVIE 294
Score = 66.6 bits (161), Expect(2) = 4e-44
Identities = 31/44 (70%), Positives = 37/44 (83%)
Frame = +3
Query: 1081 VTIRVILTLALTNHWTIQQLDVNNAFLNGDLAEEVYMQQPQGFE 1124
VTIR+IL +ALTNHW +QQ+D+NNAFL+G L EEVYM Q GFE
Sbjct: 297 VTIRLILFIALTNHWPLQQVDINNAFLHGLLTEEVYMVQLPGFE 428
>CO984748
Length = 810
Score = 174 bits (441), Expect = 2e-43
Identities = 122/310 (39%), Positives = 153/310 (49%), Gaps = 11/310 (3%)
Frame = -2
Query: 297 IQCQLCYKFGHDVWQCYHRFNPNFVPQKPPQNSPQNSQWNSSPQWHSNSQW---SSTPQW 353
IQCQ YKF HD CYHRFNPN+V Q+ QN N N S Q SN Q S Q
Sbjct: 803 IQCQXXYKFDHDAADCYHRFNPNYVAQQYNQNQSLNQHRNYSYQ-RSNCQALNNKSQSQQ 627
Query: 354 NSQWSAPVSSPWQHSSSQWPPRYPPAPSGYAPMASSPGVPYRGPAPSPMGYAQAHVAFQT 413
N+Q AP Y A+
Sbjct: 626 NAQ----------------------APQAYLANANP------------------------ 585
Query: 414 STSTPQATSPAQSWYPDSGASHHVTSESQNVHQFTPFAGQDQLFVGNGQGLPISG--IGS 471
T +Q+WY + GA+HHVT+ QN+ P +G +Q+F+GNGQGLPI+G + +
Sbjct: 584 -------TQQSQNWYINYGATHHVTASPQNLMNEVPTSGNEQVFLGNGQGLPITGSTVFN 426
Query: 472 STLSSG--LVLNNLLHVPSITKNLVSVSKFARDNGVFFEFHPYYCCVKSQASSKVLLQGS 529
S +S L LNNLLHVP ITKNLVSVS FA+DN VFFEFH +C VKSQ +LL+G+
Sbjct: 425 SPFASNVKLTLNNLLHVPHITKNLVSVS*FAKDN-VFFEFHSSHCVVKSQVDGSILLRGA 249
Query: 530 VGADGLYKFHEFKLSHLPVSLSAPVQHPVSVHTSVSS--SGHAPVEQPSVV--SSVCTAN 585
+ DGLY F S ++ + +VH S S S H S + S+ +
Sbjct: 248 LSQDGLYVFSNLLNSSFKFTVG---KAAANVHMSSLSDLSSHCNNVSTSALNTSNYSLVS 78
Query: 586 LWHFRLGHPS 595
LWH RLGHPS
Sbjct: 77 LWHNRLGHPS 48
>BF596070 similar to GP|27901698|gb gag-pol polyprotein {Vitis vinifera},
partial (34%)
Length = 407
Score = 160 bits (405), Expect = 3e-39
Identities = 72/130 (55%), Positives = 97/130 (74%), Gaps = 2/130 (1%)
Frame = -2
Query: 1025 VPLPPGREAIGCKWVFRVKHNPDGTLNKYKARLVAKGFHQQPGFDYTETFSPVIKPVTIR 1084
VPLPPG+ +GC+WV+ VK P G +++ KARLVAKG+ Q G DY +TFSPV K T+R
Sbjct: 406 VPLPPGKTPVGCRWVYTVKVGPTGEVDRLKARLVAKGYTQVYGIDYCDTFSPVAKLTTVR 227
Query: 1085 VILTLALTNHWTIQQLDVNNAFLNGDLAEEVYMQQPQGF--EHDKKLVCRLHKALYGLKQ 1142
+ L +A HW + QLD+ NAFL+GDL E++YM+QP GF + + LVC+LH++LYGLKQ
Sbjct: 226 LFLAMAAICHWPLHQLDIKNAFLHGDLEEDIYMEQPPGFVAQGEYGLVCKLHRSLYGLKQ 47
Query: 1143 APRAWFEKLT 1152
+PRAWF K +
Sbjct: 46 SPRAWFGKFS 17
>CA784773 weakly similar to GP|27901698|gb gag-pol polyprotein {Vitis
vinifera}, partial (34%)
Length = 409
Score = 157 bits (398), Expect = 2e-38
Identities = 68/128 (53%), Positives = 93/128 (72%)
Frame = +3
Query: 993 PTTVKQALAHPQWLQAMQAEFDALQKNKTWSLVPLPPGREAIGCKWVFRVKHNPDGTLNK 1052
P+T+++AL HP W QAM E AL+ N TW LVPLPPG+ +GC+WV+ VK P+G +++
Sbjct: 21 PSTIREALDHPGWRQAMVDEMQALENNGTWELVPLPPGKTTVGCRWVYTVKVGPNGKVDR 200
Query: 1053 YKARLVAKGFHQQPGFDYTETFSPVIKPVTIRVILTLALTNHWTIQQLDVNNAFLNGDLA 1112
KARLVAKG+ Q G +Y +TFSPV T+R+ L +A HW + QLD+ NAFL+GDL
Sbjct: 201 LKARLVAKGYTQVYGIEYCDTFSPVFFLTTVRLFLAMAAIRHWPLHQLDIKNAFLHGDLE 380
Query: 1113 EEVYMQQP 1120
E++YM+QP
Sbjct: 381 EDIYMEQP 404
>BU548265 homologue to GP|18000935|gb| cytochrome b {Mabuya delalandii},
partial (5%)
Length = 667
Score = 153 bits (386), Expect = 5e-37
Identities = 90/211 (42%), Positives = 125/211 (58%), Gaps = 27/211 (12%)
Frame = -3
Query: 417 TPQATSPAQSWYPDSGASHHVTSESQNVHQFTPFAGQDQLFVGNGQGLPISGIGSSTLSS 476
T A +P+QSWYPDSGASHHVT+ SQN+ Q PF Q+ +GNGQGL I+ G ST SS
Sbjct: 647 TSSAATPSQSWYPDSGASHHVTNMSQNIQQVAPFEEPIQIIIGNGQGLNINSSGLSTFSS 468
Query: 477 ------GLVLNNLLHVPSITKNLVSVSKFARDNGVFFEFHPYYCCVKSQASSKVLLQGSV 530
LVL+NLL VP+ITKNL+ VS+F +DN V+FEFH Y C KS + VLL+G V
Sbjct: 467 PINPQFSLVLSNLLFVPTITKNLIRVSQFCKDNNVYFEFHSYVCLFKS*DTKLVLLEGFV 288
Query: 531 GADGLYKFHEFKLSHLPVSLS------APVQHPVSVHTSV------------SSSGHAPV 572
G+DGLY+ + + +P S S + + + + V TS+ S+ + V
Sbjct: 287 GSDGLYQSPQL-FASMPQSKSYLKSFRSSLSNNIVVKTSIVKTTNCSTKFLFVSANNTVV 111
Query: 573 EQPSVVSSVCTA---NLWHFRLGHPSLNVLR 600
V ++ C + +WH RLGH S++ ++
Sbjct: 110 VDLVVNTTSCNSVSFPIWHSRLGHHSVDAMK 18
>AI855818 weakly similar to GP|21741393|e OSJNBb0051N19.6 {Oryza sativa
(japonica cultivar-group)}, partial (10%)
Length = 463
Score = 144 bits (362), Expect = 3e-34
Identities = 72/152 (47%), Positives = 102/152 (66%), Gaps = 2/152 (1%)
Frame = -3
Query: 1008 AMQAEFDALQKNKTWSLVPLPPGREAIGCKWVFRVKHNPDGTLNKYKARLVAKGFHQQPG 1067
AMQ E + ++N W LV P IG KWVFR K + G + + KARLVAKG++Q+ G
Sbjct: 458 AMQEELNQFERNNVWKLVEKPENYPVIGTKWVFRNKLDEHGIIIRNKARLVAKGYNQEEG 279
Query: 1068 FDYTETFSPVIKPVTIRVILTLALTNHWTIQQLDVNNAFLNGDLAEEVYMQQPQGFEHDK 1127
DY ET++PV + IR++L ++ + Q+DV +AFLNG + EEVY++QP GFE
Sbjct: 278 IDYEETYAPVARLEVIRMLLAYVSIMNFKLYQMDVKSAFLNGLIQEEVYVEQPPGFEIPD 99
Query: 1128 K--LVCRLHKALYGLKQAPRAWFEKLTSALVQ 1157
K V +L KALYGLKQAPRAW+E++++ L++
Sbjct: 98 KPTHVYKLQKALYGLKQAPRAWYERISNFLLE 3
>BM892610
Length = 421
Score = 141 bits (356), Expect = 1e-33
Identities = 60/97 (61%), Positives = 80/97 (81%)
Frame = -3
Query: 778 VYLINRLPTSSLQFDIPLYKLFKQIPDYTFLKVFGCTCFPLMRPYNSHKLDFRSQECVFL 837
VYLINRLPT+SL+F +P +F ++P+Y FL+ FGC+C+P +RPYN HKLDFRS EC+FL
Sbjct: 416 VYLINRLPTTSLKFQVPC-TVFNKLPNYNFLRNFGCSCYPFLRPYNKHKLDFRSHECLFL 240
Query: 838 GYSPSHRGYKCLASSGRIYISKDVIFDEHTFPYPKLF 874
GYS H+GYKCL+ +G++Y+SKDVIF+E +PY LF
Sbjct: 239 GYSTPHKGYKCLSPNGKLYVSKDVIFNELKYPYSDLF 129
>BF595216
Length = 421
Score = 79.3 bits (194), Expect(2) = 2e-31
Identities = 37/50 (74%), Positives = 44/50 (88%)
Frame = +1
Query: 1043 KHNPDGTLNKYKARLVAKGFHQQPGFDYTETFSPVIKPVTIRVILTLALT 1092
K N DGT+NKYKA+LVAKGFHQQ G DY ETFS VIKP+T+R++LTLA+T
Sbjct: 124 KENSDGTVNKYKAKLVAKGFHQQYGTDYIETFSLVIKPITMRLLLTLAVT 273
Score = 76.6 bits (187), Expect(2) = 2e-31
Identities = 37/49 (75%), Positives = 40/49 (81%), Gaps = 1/49 (2%)
Frame = +3
Query: 1091 LTNHWTIQQLDVNNAFLNGDLAEEVYMQQPQGFEH-DKKLVCRLHKALY 1138
LT HW IQQLDVNNAFLNG L EEVYMQQP GF+ K LVC+LHKA+Y
Sbjct: 273 LTYHWPIQQLDVNNAFLNGILQEEVYMQQPPGFDSTTKSLVCKLHKAIY 419
Score = 61.6 bits (148), Expect = 2e-09
Identities = 25/40 (62%), Positives = 34/40 (84%)
Frame = +2
Query: 647 YSDLWGPSPSPSSSGYLYYITFIDAYSRYTWIYLLKAKSE 686
+SDL P+P SS+GY YY+TF+DA++RYTWI+LL+ KSE
Sbjct: 5 FSDLCSPAPFVSSTGYSYYVTFVDAFTRYTWIFLLRHKSE 124
>BQ612840
Length = 441
Score = 120 bits (300), Expect(2) = 3e-29
Identities = 63/112 (56%), Positives = 74/112 (65%), Gaps = 6/112 (5%)
Frame = +2
Query: 419 QATSPAQSWYPDSGASHHVTSESQNVHQFTPFAGQDQLFVGNGQGLPISGIGSSTLSSG- 477
Q S + W PDSGAS H ESQN+ Q PF G D++F+GNGQ L I+ GSS+ S
Sbjct: 44 QPNSNSVIWIPDSGASCHFIGESQNIQQIEPFEGIDRIFIGNGQDLKITSSGSSSFISPY 223
Query: 478 -----LVLNNLLHVPSITKNLVSVSKFARDNGVFFEFHPYYCCVKSQASSKV 524
L LN LHVP ITKNLVSVS+FARDNG FEFH YC VKSQ +++V
Sbjct: 224 NSKVFLSLNRFLHVPYITKNLVSVSQFARDNGFCFEFHSNYCAVKSQVTNEV 379
Score = 28.5 bits (62), Expect(2) = 3e-29
Identities = 11/20 (55%), Positives = 13/20 (65%)
Frame = +1
Query: 526 LQGSVGADGLYKFHEFKLSH 545
LQG+VG DGLY F + H
Sbjct: 379 LQGTVGPDGLYSFSNIQFQH 438
>TC235104 weakly similar to UP|Q850H8 (Q850H8) Gag-pol polyprotein
(Fragment), partial (28%)
Length = 865
Score = 122 bits (307), Expect = 7e-28
Identities = 92/312 (29%), Positives = 146/312 (46%), Gaps = 2/312 (0%)
Frame = +2
Query: 458 VGNGQGLPISGIGSSTLSSGLVLNNLLHVPSITKNLVSVSKFARDNGVFFEFHPYYCCVK 517
+ +G + +GIG + +S L LN+++ + N+ S+S+ R C V
Sbjct: 26 LADGSRVVATGIGHVSPTSSLSLNSVVFILGCPFNITSLSQLTRFRN---------CSVT 178
Query: 518 SQASSKVLLQGSVGADGLYKFHEFKLSHLPVSLSAPVQHPVSVHTSVSSSGHAPVEQPSV 577
A+S V+ + G L +L +LS V
Sbjct: 179 FDANSFVIQECGTGWTIGVGIESHGLYYLKPNLSW------------------------V 286
Query: 578 VSSVCTANLWHFRLGHPSLNVLRKALQFCKIPVSNKMDVDFCKACVMGKSHRLPSSLSTT 637
S+V + L H RLGHP L+ L+ +P K+ FC++C +GK R +
Sbjct: 287 CSAVTSPKLLHERLGHPHLSKLK-----IMVPSLEKIKDLFCESCQLGKHVRSSXRHVES 451
Query: 638 EYTSPLELVYSDLWGPSPSPSSSGYLYYITFIDAYSRYTWIYLLKAKSEAISAFHQFKAS 697
SP +++ D+WGP+ SS Y Y++TFID +S+ T ++L+K +SE +S F
Sbjct: 452 RVDSPFLVIHXDIWGPN-RVSSMSYRYFVTFIDEFSQCTRVFLMKERSEILS-FLTSVNK 625
Query: 698 SELQLNAKIKALQTDWGGEF--RVFPNFLKDFGIHHRLICPHTHHQNGVVERKHRHIVDL 755
+ Q IK L++D E+ V F GI H+ CPHT QN + ERK+RH+V+
Sbjct: 626 IKTQFGKTIKILRSDNAKEYFSSVISPFXSAQGILHQFSCPHTPQQNDIAERKNRHLVET 805
Query: 756 GLSLLAHASIPL 767
+LL HA+ P+
Sbjct: 806 ARTLLLHANEPI 841
>TC224181
Length = 597
Score = 116 bits (290), Expect(2) = 2e-27
Identities = 71/149 (47%), Positives = 92/149 (61%), Gaps = 10/149 (6%)
Frame = +2
Query: 450 FAGQDQLFVGNGQGLPISGIGSSTLSSGL------VLNNLLHVPSITKNLVSVSKFARDN 503
F G DQ+F+GN QGL I G S S L VL NLLHVPSITKNL+SVS+FARDN
Sbjct: 152 FDGPDQIFIGNSQGLHIKGSSCSFFKSPLNSKFSFVLKNLLHVPSITKNLLSVSQFARDN 331
Query: 504 GVFFEFHPYYCCVKSQASSKVLLQGSVGADGLYKFHEFKL--SHLPVSLSAPVQHPVSVH 561
V+FEFHPY VKSQA+ L+QG++GA+GL+ F L S +S S+ + +
Sbjct: 332 SVYFEFHPYTFVVKSQATHGPLIQGTIGANGLHSFTHVNLHPSSALLSSSSSISPSSLSN 511
Query: 562 TSVSSSG--HAPVEQPSVVSSVCTANLWH 588
S++SS H S+V S T+++WH
Sbjct: 512 KSLTSSPFVHTINSDSSIVPS--TSSIWH 592
Score = 26.2 bits (56), Expect(2) = 2e-27
Identities = 10/20 (50%), Positives = 14/20 (70%)
Frame = +1
Query: 435 HHVTSESQNVHQFTPFAGQD 454
HHVT SQN+HQ ++G +
Sbjct: 112 HHVTGASQNIHQAF*WSGSN 171
>BM307983
Length = 406
Score = 120 bits (301), Expect = 4e-27
Identities = 63/134 (47%), Positives = 84/134 (62%), Gaps = 3/134 (2%)
Frame = +2
Query: 1034 IGCKWVFRVKHNPDGTLNKYKARLVAKGFHQQPGFDYTETFSPVIKPVTIRVILTLALTN 1093
+GC+W++ VK+ D TL++YKARLVAKG+ Q G DY ETF+ K +
Sbjct: 2 VGCRWIYTVKY*ADDTLDRYKARLVAKGYIQTYGIDYEETFAQWQK*IQSGSSSP*QQAQ 181
Query: 1094 H-WTIQQLDVNNAFLNGDLAEEVYMQQPQGF--EHDKKLVCRLHKALYGLKQAPRAWFEK 1150
W + Q DV NAFL+G L EEVYM+ P G+ + VCRL KALYGLKQ+PRAWF +
Sbjct: 182 FGWEMHQFDVKNAFLHGSLEEEVYMEIPPGYGASNGGNKVCRLKKALYGLKQSPRAWFGR 361
Query: 1151 LTSALVQFGFVASK 1164
T A++ G+ S+
Sbjct: 362 FTQAMLSLGYKQSQ 403
>CO983154
Length = 568
Score = 117 bits (293), Expect = 3e-26
Identities = 74/191 (38%), Positives = 106/191 (54%), Gaps = 2/191 (1%)
Frame = +3
Query: 737 HTHHQNGVVERKHRHIVDLGLSLLAHASIPLTYWDFAFSTAVYLINRLPTSSLQFDIPLY 796
HT QNG+ ERK+RH+++ SL+ + ++P+ +W A T+ +LINR+P+SSL+ IP
Sbjct: 3 HTPQQNGIAERKNRHLLETARSLMLNLNVPIHHWGDAVLTSCFLINRMPSSSLENQIPHS 182
Query: 797 KLFKQIP-DYTFLKVFGCTCFPLMRPYNSHKLDFRSQECVFLGYSPSHRGYKCLA-SSGR 854
+F P + KVFGCTCF KL RS +CVFLGYS +GYKC + + R
Sbjct: 183 LVFPHDPLFHVSPKVFGCTCFVHDLSPGLDKLSARSVKCVFLGYSRLQKGYKCYSPTMRR 362
Query: 855 IYISKDVIFDEHTFPYPKLFAQNSNSPSLSPAVVLPSLPSVEHFSQPAIAQPSSSLPSPF 914
Y+S DV F E T P +S SL + +PS + + Q PSSS S
Sbjct: 363 YYMSADVTFFEDT---PFFSPSVDHSSSLQEVLPIPSPYPLXNSGQNVSIVPSSSPNSLE 533
Query: 915 LPSPIASSDQN 925
+ P ++DQ+
Sbjct: 534 VILPPLTTDQH 566
>AI959950
Length = 466
Score = 115 bits (288), Expect = 1e-25
Identities = 59/139 (42%), Positives = 90/139 (64%)
Frame = -1
Query: 1004 QWLQAMQAEFDALQKNKTWSLVPLPPGREAIGCKWVFRVKHNPDGTLNKYKARLVAKGFH 1063
Q ++AMQ E D QKN LV LP ++ +G KW+F K + DG + +YKARLVAKG+
Sbjct: 403 QLMKAMQEELDQFQKNNV*KLVKLPKRKKVVGVKWIFCNKLDEDGKVVRYKARLVAKGYS 224
Query: 1064 QQPGFDYTETFSPVIKPVTIRVILTLALTNHWTIQQLDVNNAFLNGDLAEEVYMQQPQGF 1123
QQ G DY +TF+ V + I ++L+ A ++ + Q+DV +AFLNG + +EVY++QP GF
Sbjct: 223 QQEGIDYPKTFALVARLEVICILLSFATYSNMKLYQMDVKSAFLNGLIQKEVYVEQPPGF 44
Query: 1124 EHDKKLVCRLHKALYGLKQ 1142
E++ LH+ ++ L +
Sbjct: 43 ENE-----TLHQHVFKLNK 2
>BG508014 GP|15128578|db prolactin receptor {Paralichthys olivaceus}, partial
(2%)
Length = 450
Score = 112 bits (279), Expect = 1e-24
Identities = 58/145 (40%), Positives = 89/145 (61%)
Frame = +1
Query: 7 PPPPPPASPLPPHRFKLPSLSTYTISEKLSDRNFLLWKQQVEPIIKAHNLHVLVQDPEIP 66
PP PP+S P K S ++IS KL N+LLW QQVE +IK+ +LH + +P IP
Sbjct: 16 PPLFPPSSSSSPTSLKQFS---HSISHKLHVTNYLLWLQQVELVIKSRHLHHFLVNPLIP 186
Query: 67 PRFVTERDRVAGTLNPAYSVWESKDQYLLSWLQSSLSTPVLSRMIGCAHSFQLWEKIHSF 126
++ +D + T Y W+ +DQ+ L+WL+S++ +L+ +IGC S+QLW+++
Sbjct: 187 KKYALVKDCDSDTTTEVYHGWKEQDQFFLAWLRSTIYGDMLTHVIGCKSSWQLWDRLQLH 366
Query: 127 FHNQTRAKSTQLRTELRSETLGNRT 151
F + TRA+ QLR ELR + GNR+
Sbjct: 367 FQSLTRARVRQLRNELRCLSHGNRS 441
>BI427153
Length = 422
Score = 111 bits (278), Expect = 2e-24
Identities = 60/139 (43%), Positives = 83/139 (59%), Gaps = 2/139 (1%)
Frame = +1
Query: 731 HRLICPHTHHQNGVVERKHRHIVDLGLSLLAHASIPLTYWDFAFSTAVYLINRLPTSSLQ 790
H+ CPHT QNG+ ERK+ H+++ SL+ ++++P +W A TA +LINR+P+SSL+
Sbjct: 1 HQSTCPHTPQQNGIAERKNHHLLETARSLMLNSNVPTHHWGDAVLTACFLINRMPSSSLE 180
Query: 791 FDIPLYKLF-KQIPDYTFLKVFGCTCFPLMRPYNSHKLDFRSQECVFLGYSPSHRGYKC- 848
IP +F + Y KVFGCTCF KL RS +CVFLGYS +GY C
Sbjct: 181 NQIPHSIVFPNDLLFYVSPKVFGCTCFVHDLSPGLDKLSARSVKCVFLGYSRLQKGYTCY 360
Query: 849 LASSGRIYISKDVIFDEHT 867
+ R Y+S +V F E T
Sbjct: 361 FPNMRRYYMSANVTFFEDT 417
>BF068957 weakly similar to PIR|B85188|B85 retrotransposon like protein
[imported] - Arabidopsis thaliana, partial (3%)
Length = 420
Score = 110 bits (274), Expect = 5e-24
Identities = 53/126 (42%), Positives = 85/126 (67%)
Frame = -2
Query: 98 LQSSLSTPVLSRMIGCAHSFQLWEKIHSFFHNQTRAKSTQLRTELRSETLGNRTISEFLL 157
LQS+LS VLSR++ HS+Q+W+KIH F T+ ++ QL T +R+ TL +TI E L
Sbjct: 419 LQSTLSKSVLSRVLASDHSYQVWDKIHEHFSLHTKTRAPQLHTAMRAVTLDGKTIEE*L* 240
Query: 158 QIKSIIDSLFAIGDPISPREHLDVILQGLPSDYEDLITLISTRFEEHSIDELEALLLAKE 217
+IK ++ L ++G P+ +E+++ +L+GLPSDY ++++I ++ SI E+EALL E
Sbjct: 239 KIKGFVNELASVGVPVHHKEYVEALLEGLPSDYASVVSVIESKKRTPSIAEIEALLYRHE 60
Query: 218 ARLGKY 223
RL +Y
Sbjct: 59 TRLMRY 42
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.317 0.132 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 65,742,711
Number of Sequences: 63676
Number of extensions: 1272318
Number of successful extensions: 26057
Number of sequences better than 10.0: 891
Number of HSP's better than 10.0 without gapping: 12904
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 21707
length of query: 1182
length of database: 12,639,632
effective HSP length: 108
effective length of query: 1074
effective length of database: 5,762,624
effective search space: 6189058176
effective search space used: 6189058176
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 64 (29.3 bits)
Lotus: description of TM0336.1


