

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
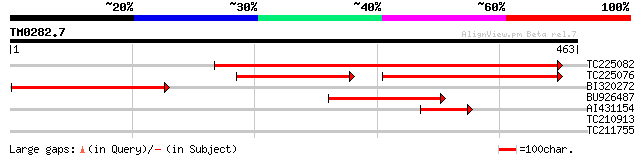
Query= TM0282.7
(463 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC225082 similar to UP|Q9M6U1 (Q9M6U1) Homogentisate 1,2-dioxyge... 565 e-161
TC225076 similar to UP|Q9M6U1 (Q9M6U1) Homogentisate 1,2-dioxyge... 287 e-128
BI320272 similar to GP|8131905|gb| homogentisate 1 2-dioxygenase... 241 6e-64
BU926487 homologue to GP|8131905|gb|A homogentisate 1 2-dioxygen... 204 9e-53
AI431154 64 2e-10
TC210913 similar to UP|O13524 (O13524) B2-aldehyde-forming enzym... 28 6.6
TC211755 similar to UP|Q9SAK1 (Q9SAK1) T8K14.10, partial (22%) 28 8.6
>TC225082 similar to UP|Q9M6U1 (Q9M6U1) Homogentisate 1,2-dioxygenase
(Fragment) , partial (56%)
Length = 1234
Score = 565 bits (1455), Expect = e-161
Identities = 262/284 (92%), Positives = 272/284 (95%)
Frame = +3
Query: 168 LFITTECGRLKVSPGEIAMLPQGFRFTVDLPDGPSRGYVAEIFGTHFQLPDLGPIGANGL 227
L +TTECGRLKVSPGEIA+LPQGFRF+V+LPDGPSRGYVAEIFGTHFQLPDLGPIGANGL
Sbjct: 21 LLVTTECGRLKVSPGEIAILPQGFRFSVNLPDGPSRGYVAEIFGTHFQLPDLGPIGANGL 200
Query: 228 AAPRDFLVPTAWFEDKSYPGYTIVQKFGGELFSAVQDFSPFNVVAWRGNYVPYKYDLSKF 287
A+PRDFLVPTAWFEDKSYPGYTIVQKFGGELF AVQDFSPFNVVAW GNYVPYKYDL+KF
Sbjct: 201 ASPRDFLVPTAWFEDKSYPGYTIVQKFGGELFDAVQDFSPFNVVAWHGNYVPYKYDLNKF 380
Query: 288 CPYNTVLFDHSDPSINTVLTAPTDKPGVALLDFVIFPPRWLVAEHTFRPPYYHRNCMSEF 347
CPYNTVLFDHSDPSINTVLTAPTDKPGVALLDFVIFPPRWLVAEHTFRPPYYHRNCMSEF
Sbjct: 381 CPYNTVLFDHSDPSINTVLTAPTDKPGVALLDFVIFPPRWLVAEHTFRPPYYHRNCMSEF 560
Query: 348 MGLIHGGYEAKVDGFLPGGASLHSCMTPHGPDTKSYEATIARGNDVGPNKITDTMAFMFE 407
MGLIHGGYEAK DGFLPGGASLHSCMTPHGPDTKSYEATIARGNDVGP KITDTMAFMFE
Sbjct: 561 MGLIHGGYEAKADGFLPGGASLHSCMTPHGPDTKSYEATIARGNDVGPCKITDTMAFMFE 740
Query: 408 SCLIPRISRWALESPFLDHDYYQCWIGLRSHFNVTEISPRTPTL 451
S LIPRIS+WA ESPFLD DYYQCWIGL+SHF VT+ SP P+L
Sbjct: 741 SSLIPRISQWASESPFLDQDYYQCWIGLKSHFAVTKTSPENPSL 872
>TC225076 similar to UP|Q9M6U1 (Q9M6U1) Homogentisate 1,2-dioxygenase
(Fragment) , partial (48%)
Length = 1391
Score = 287 bits (735), Expect(2) = e-128
Identities = 133/147 (90%), Positives = 136/147 (92%)
Frame = +1
Query: 305 VLTAPTDKPGVALLDFVIFPPRWLVAEHTFRPPYYHRNCMSEFMGLIHGGYEAKVDGFLP 364
VLTAPTDKPGVALLDFVIFPPRWLVAEHTFRPPYYHRNCMSEFMGLIHGGYEAK DGFLP
Sbjct: 289 VLTAPTDKPGVALLDFVIFPPRWLVAEHTFRPPYYHRNCMSEFMGLIHGGYEAKADGFLP 468
Query: 365 GGASLHSCMTPHGPDTKSYEATIARGNDVGPNKITDTMAFMFESCLIPRISRWALESPFL 424
GGASLH+CMTPHGPDTKSYEATIARGND GP KITDTMAFMFES LIPRIS+WALESPFL
Sbjct: 469 GGASLHNCMTPHGPDTKSYEATIARGNDGGPCKITDTMAFMFESSLIPRISQWALESPFL 648
Query: 425 DHDYYQCWIGLRSHFNVTEISPRTPTL 451
D DYYQCWIGL+SHF VTE SP L
Sbjct: 649 DQDYYQCWIGLKSHFTVTETSPENTNL 729
Score = 190 bits (482), Expect(2) = e-128
Identities = 86/96 (89%), Positives = 92/96 (95%)
Frame = +2
Query: 186 MLPQGFRFTVDLPDGPSRGYVAEIFGTHFQLPDLGPIGANGLAAPRDFLVPTAWFEDKSY 245
++P GFRF+V+L DGPSRGYVAEIFGTHFQLPDLGPIGANGLA+PRDFLVP+AWFEDKSY
Sbjct: 2 IIPHGFRFSVNLSDGPSRGYVAEIFGTHFQLPDLGPIGANGLASPRDFLVPSAWFEDKSY 181
Query: 246 PGYTIVQKFGGELFSAVQDFSPFNVVAWRGNYVPYK 281
PGYTIVQKFGGELF AVQDFSPFNVVAW GNYVPYK
Sbjct: 182 PGYTIVQKFGGELFDAVQDFSPFNVVAWHGNYVPYK 289
>BI320272 similar to GP|8131905|gb| homogentisate 1 2-dioxygenase
{Lycopersicon esculentum}, partial (24%)
Length = 389
Score = 241 bits (614), Expect = 6e-64
Identities = 113/129 (87%), Positives = 120/129 (92%)
Frame = +2
Query: 2 EHSTAGDDFNYLSGFGNHFSSEALAGALPEAQNSPLICPYGLYAEQISGTSFTSPRNLNL 61
E+ G +F YLSGFGNHFSSEALAGALP AQNSPL+CPYGLYAEQISGTSFTSPRN NL
Sbjct: 2 ENPIDGGEFVYLSGFGNHFSSEALAGALPVAQNSPLVCPYGLYAEQISGTSFTSPRNRNL 181
Query: 62 FSWFYRVKPSVTHEPFKPRVPCNGRILSEFNHSNSSTNPTQLRWKPMDSPDSPTDFIDGL 121
FSWFYR+KPSVTHEPFKPRVP NGRILSEFN+SNSS NPTQLRWKP+D+PDSPTDFIDGL
Sbjct: 182 FSWFYRIKPSVTHEPFKPRVPGNGRILSEFNNSNSSANPTQLRWKPLDAPDSPTDFIDGL 361
Query: 122 STVCGSGSS 130
STVCGSGSS
Sbjct: 362 STVCGSGSS 388
>BU926487 homologue to GP|8131905|gb|A homogentisate 1 2-dioxygenase
{Lycopersicon esculentum}, partial (20%)
Length = 445
Score = 204 bits (518), Expect = 9e-53
Identities = 93/96 (96%), Positives = 93/96 (96%)
Frame = +3
Query: 261 AVQDFSPFNVVAWRGNYVPYKYDLSKFCPYNTVLFDHSDPSINTVLTAPTDKPGVALLDF 320
AVQDFSPFNVVAW GNYVP KYDLSKFCPYNTVLFDHSDPSINTVLTAPTDKPGVALLDF
Sbjct: 117 AVQDFSPFNVVAWHGNYVP*KYDLSKFCPYNTVLFDHSDPSINTVLTAPTDKPGVALLDF 296
Query: 321 VIFPPRWLVAEHTFRPPYYHRNCMSEFMGLIHGGYE 356
VIFPPRWLVAEHTF PPYYHRNCMSEFMGLIHGGYE
Sbjct: 297 VIFPPRWLVAEHTFLPPYYHRNCMSEFMGLIHGGYE 404
Score = 30.4 bits (67), Expect = 1.7
Identities = 11/14 (78%), Positives = 13/14 (92%)
Frame = +2
Query: 141 YTANKSMNNCAFCN 154
Y ANKSM+NCAFC+
Sbjct: 80 YNANKSMDNCAFCS 121
>AI431154
Length = 130
Score = 63.5 bits (153), Expect = 2e-10
Identities = 28/43 (65%), Positives = 31/43 (71%)
Frame = +2
Query: 336 PPYYHRNCMSEFMGLIHGGYEAKVDGFLPGGASLHSCMTPHGP 378
PPY +R EFMG I GGY AK DGFLP GA LHSC+T +GP
Sbjct: 2 PPYDYRKWAKEFMGPIQGGY*AKADGFLPAGARLHSCLTTYGP 130
>TC210913 similar to UP|O13524 (O13524) B2-aldehyde-forming enzyme, partial
(10%)
Length = 593
Score = 28.5 bits (62), Expect = 6.6
Identities = 17/45 (37%), Positives = 23/45 (50%)
Frame = +1
Query: 274 RGNYVPYKYDLSKFCPYNTVLFDHSDPSINTVLTAPTDKPGVALL 318
RG YV Y +D P N V +H NT++ A D PGV ++
Sbjct: 394 RGGYVTYVHDSCM*VPGNAV--NHLY*LQNTIIFAVIDPPGVVIM 522
>TC211755 similar to UP|Q9SAK1 (Q9SAK1) T8K14.10, partial (22%)
Length = 1127
Score = 28.1 bits (61), Expect = 8.6
Identities = 15/48 (31%), Positives = 27/48 (56%), Gaps = 1/48 (2%)
Frame = +1
Query: 300 PSINTVLTAPT-DKPGVALLDFVIFPPRWLVAEHTFRPPYYHRNCMSE 346
PS++ +AP+ + P + L V+FPP + H +PP Y C+++
Sbjct: 562 PSLSPPKSAPSLNPPPLVYLPPVVFPPPPSPSTHRKKPPQYALWCVAK 705
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.321 0.139 0.446
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 24,194,146
Number of Sequences: 63676
Number of extensions: 390720
Number of successful extensions: 1708
Number of sequences better than 10.0: 14
Number of HSP's better than 10.0 without gapping: 1681
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1707
length of query: 463
length of database: 12,639,632
effective HSP length: 101
effective length of query: 362
effective length of database: 6,208,356
effective search space: 2247424872
effective search space used: 2247424872
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0282.7


