

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
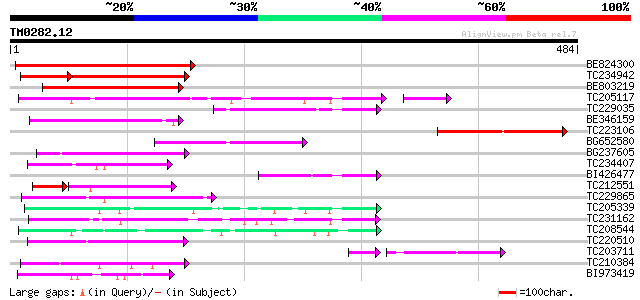
Query= TM0282.12
(484 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BE824300 156 2e-38
TC234942 weakly similar to UP|Q9LVW3 (Q9LVW3) Flavonol 3-O-gluco... 99 2e-34
BE803219 130 1e-30
TC205117 weakly similar to UP|Q940V3 (Q940V3) At2g22590/T9I22.3,... 115 6e-29
TC229035 weakly similar to UP|Q8LP23 (Q8LP23) Rhamnosyl transfer... 112 5e-25
BE346159 similar to GP|10177261|dbj flavonol 3-O-glucosyltransfe... 98 9e-21
TC223106 weakly similar to UP|Q9T081 (Q9T081) UDP rhamnose--anth... 96 5e-20
BG652580 similar to GP|22759895|dbj rhamnosyl transferase {Niere... 94 1e-19
BG237605 weakly similar to GP|8809576|dbj| flavonol 3-O-glucosyl... 89 6e-18
TC234407 weakly similar to GB|AAO63454.1|28951061|BT005390 At5g4... 75 7e-14
BI426477 weakly similar to GP|10177261|dbj flavonol 3-O-glucosyl... 75 9e-14
TC212551 similar to UP|Q9LSM0 (Q9LSM0) Anthocyanidin-3-glucoside... 46 1e-11
TC229865 weakly similar to UP|P93789 (P93789) UDP-glucose glucos... 56 3e-08
TC205339 weakly similar to UP|Q6QDB6 (Q6QDB6) UDP-glucose glucos... 54 1e-07
TC231162 weakly similar to UP|Q9FUJ6 (Q9FUJ6) UDP-glucosyltransf... 54 2e-07
TC208544 weakly similar to PIR|D96766|D96766 protein glucosyltra... 52 6e-07
TC220510 similar to UP|Q7XZD0 (Q7XZD0) Isoflavonoid glucosyltran... 48 9e-06
TC203711 similar to UP|Q8S9A4 (Q8S9A4) Glucosyltransferase-5, pa... 42 1e-05
TC210384 weakly similar to UP|Q8VZF9 (Q8VZF9) AT5g12890/T24H18_6... 45 9e-05
BI973419 44 1e-04
>BE824300
Length = 614
Score = 156 bits (394), Expect = 2e-38
Identities = 70/153 (45%), Positives = 100/153 (64%)
Frame = +3
Query: 6 SKNTSSSNSKLHIAMFPWFAMGHMTPYVHLANELAKRGHKITFLLPNKARLALHHLNRYP 65
S+ T+ S LHIAMFPWFAMGH+TPY+HL+N+LAKRGH+I+F +P + L N +P
Sbjct: 153 SRQTNMDASSLHIAMFPWFAMGHLTPYLHLSNKLAKRGHRISFFIPKRTPHKLEQFNLFP 332
Query: 66 NLITFHIITVPNVDGLPLGTETASEIPLSLNHLLVVAMDKTRNQVEQVMKSKKPHFVLFD 125
+LITF I VP+V+GLP G ET S++ SL L++ AMD+T +E ++ P V FD
Sbjct: 333 HLITFFPINVPHVEGLPHGAETTSDVSFSLAPLIMTAMDRTEKDIELLLIELMPQIVFFD 512
Query: 126 VAYWVPEAARKLGIKTICYHVVSAMSMAMSFVP 158
YW+P R+ GIK+ Y ++ +++ P
Sbjct: 513 FTYWLPNLTRRXGIKSF*YLIIGXXTVSYXXXP 611
>TC234942 weakly similar to UP|Q9LVW3 (Q9LVW3) Flavonol
3-O-glucosyltransferase-like, partial (8%)
Length = 439
Score = 98.6 bits (244), Expect(2) = 2e-34
Identities = 45/103 (43%), Positives = 67/103 (64%)
Frame = +1
Query: 51 PNKARLALHHLNRYPNLITFHIITVPNVDGLPLGTETASEIPLSLNHLLVVAMDKTRNQV 110
P K + L HLN +P+LITF I VP+V+GLP ET S++P SL L+ AMD+T +
Sbjct: 130 PKKTQTKLQHLNLHPHLITFVPIKVPHVNGLPHDAETTSDVPFSLFPLIAEAMDRTEKDI 309
Query: 111 EQVMKSKKPHFVLFDVAYWVPEAARKLGIKTICYHVVSAMSMA 153
E +++ KP V FD +W+P R+LGIK++ Y +++ +S A
Sbjct: 310 EILLRELKPQIVFFDFQHWLPNLTRRLGIKSVMYVIINPLSTA 438
Score = 65.9 bits (159), Expect(2) = 2e-34
Identities = 26/44 (59%), Positives = 39/44 (88%)
Frame = +3
Query: 10 SSSNSKLHIAMFPWFAMGHMTPYVHLANELAKRGHKITFLLPNK 53
+++ + LHIA+FPWFAMGH+TP +HL+N+LA+RGH+I+F+ P K
Sbjct: 6 TANAAPLHIAIFPWFAMGHLTPSLHLSNKLAQRGHRISFIGPKK 137
>BE803219
Length = 428
Score = 130 bits (328), Expect = 1e-30
Identities = 62/121 (51%), Positives = 86/121 (70%), Gaps = 1/121 (0%)
Frame = +1
Query: 29 MTPYVHLANELAKRGHKITFLLPNKARLALHHLNRYPNLITFHIITVPNVDGLPLGTETA 88
+TP++HLAN+LAKRGHKI+F +P + + L LN +PNLITF I VP+VDGLP ET
Sbjct: 1 LTPFLHLANKLAKRGHKISFFIPRRTQAKLEDLNLHPNLITFVPINVPHVDGLPYDAETT 180
Query: 89 SEIPLSLNHLLVVAMDKTRNQVEQVMKSKKPHFVLFDVA-YWVPEAARKLGIKTICYHVV 147
S++P SL L+ AMD T +E ++ KPH VLFD + YW+P AR++GIK++ Y ++
Sbjct: 181 SDVPSSLFPLIGTAMDLTEKNIELLLLDLKPHIVLFDFSTYWLPNLARRIGIKSLQYWII 360
Query: 148 S 148
S
Sbjct: 361 S 363
>TC205117 weakly similar to UP|Q940V3 (Q940V3) At2g22590/T9I22.3, partial
(17%)
Length = 1693
Score = 115 bits (287), Expect(2) = 6e-29
Identities = 93/330 (28%), Positives = 145/330 (43%), Gaps = 16/330 (4%)
Frame = +2
Query: 8 NTSSSNSKLHIAMFPWFAMGHMTPYVHLANELAKRGHKITFLLP----NKARLALHHLNR 63
N S++ LH+AM PW AMGH+ PY +A LA++GH +TF+ ++ HL
Sbjct: 92 NGKSNDKPLHVAMLPWLAMGHIYPYFEVAKILAQKGHFVTFINSPKNIDRMPKTPKHLEP 271
Query: 64 YPNLITFHIITVPNVDGLPLGTETASEIPLSLNHLLVVAMDKTRNQVEQVMKSKKPHFVL 123
+ L+ + +P ++ LP G E+ +IP N L A + + V +++K+ P +VL
Sbjct: 272 FIKLVK---LPLPKIEHLPEGAESTMDIPSKKNCFLKKAYEGLQYAVSKLLKTSNPDWVL 442
Query: 124 FD-VAYWVPEAARKLGIKTICYHVVSAMSMAMSFVPAKIIPKDRPLTVEDMSQPPPGYPS 182
+D A WV A+ I Y++ A + F P K KD L + PP P
Sbjct: 443 YDFAAAWVIPIAKSYNIPCAHYNITPAFNKVF-FDPPKDKMKDYSLA--SICGPPTWLPF 613
Query: 183 SSNSKL----VLRRYEAEQQMFASIPFGEGGVTLYDRTTTGLRECDAIAIKTTTEVEGCF 238
++ + LR YE + E G CD ++T+ E+EG +
Sbjct: 614 TTTIHIRPYEFLRAYEGTKD-------EETGERASFDLNKAYSSCDLFLLRTSRELEGDW 772
Query: 239 CDYIAIQYHKNV----LLTGPVLSEEAEGEELEERWAK---WLDGLGLGLGLGLGLGPVV 291
DY+A Y V LL + + E E+ W + WLD VV
Sbjct: 773 LDYLAGNYKVPVVPVGLLPPSMQIRDVEEEDNNPDWVRIKDWLD--------TQESSSVV 928
Query: 292 YCAFGSQVILGKAQFQEILLGLEISGFSIF 321
Y FGS++ L + E+ G+E+S F
Sbjct: 929 YIGFGSELKLSQEDLTELAHGIELSNLPFF 1018
Score = 30.8 bits (68), Expect(2) = 6e-29
Identities = 16/41 (39%), Positives = 23/41 (56%)
Frame = +1
Query: 337 VAGGV*REGEGKGNGFERVGPTVVDLEAWFSGVLCESLWAW 377
VA V*R+ +G + E +G DL +W + + ESLW W
Sbjct: 1054 VARRV*RKNQGTWHCLEDLGTPA*DLGSWSNWRVHESLWLW 1176
>TC229035 weakly similar to UP|Q8LP23 (Q8LP23) Rhamnosyl transferase, partial
(36%)
Length = 638
Score = 112 bits (279), Expect = 5e-25
Identities = 60/143 (41%), Positives = 84/143 (57%)
Frame = +3
Query: 175 QPPPGYPSSSNSKLVLRRYEAEQQMFASIPFGEGGVTLYDRTTTGLRECDAIAIKTTTEV 234
+PPPGYP +SN + L+ +EA MF FGE +T Y+R L EC I KT E+
Sbjct: 9 KPPPGYPQNSN--ISLKAFEAMDFMFLFTRFGEKNLTGYERVLQSLGECSFIVFKTCKEI 182
Query: 235 EGCFCDYIAIQYHKNVLLTGPVLSEEAEGEELEERWAKWLDGLGLGLGLGLGLGPVVYCA 294
EG + DYI Q+ K VLL+GP++ E + + LEE+W+KWLD G V+ C+
Sbjct: 183 EGPYLDYIETQFRKPVLLSGPLVPEPST-DVLEEKWSKWLD--------GFPAKSVILCS 335
Query: 295 FGSQVILGKAQFQEILLGLEISG 317
FGS+ L Q +E+ GLE++G
Sbjct: 336 FGSETFLSDYQIKELASGLELTG 404
>BE346159 similar to GP|10177261|dbj flavonol 3-O-glucosyltransferase-like
protein {Arabidopsis thaliana}, partial (5%)
Length = 415
Score = 97.8 bits (242), Expect = 9e-21
Identities = 52/135 (38%), Positives = 77/135 (56%), Gaps = 4/135 (2%)
Frame = +2
Query: 18 IAMFPWFAMGHMTPYVHLANELAKRGHKITFLLPNKARLALHHLNRYPNLITFHIITVPN 77
I M W AMGH+TP++HLAN A++GH+++ +P + L LN PNLITF ++VP+
Sbjct: 2 ITMNAWLAMGHLTPFLHLANAQAQKGHRVSLFIPRATQAKLEDLNLEPNLITFVPMSVPH 181
Query: 78 VDGLPLGTETASEIPLSLNHLLVVAMDKTRNQVEQVMKSKKPHFVLFDVAYWVPEAARKL 137
DG P T S P S+ L+ AMD T +E ++ KPH D + + A KL
Sbjct: 182 GDGHPYDA*TTSHAPSSMCPLIATAMDLTHKNIELILLDLKPHIARCDTLHIL---ATKL 352
Query: 138 G----IKTICYHVVS 148
G IK++ Y +++
Sbjct: 353 GSGI*IKSLQYWIIN 397
>TC223106 weakly similar to UP|Q9T081 (Q9T081) UDP
rhamnose--anthocyanidin-3-glucoside
rhamnosyltransferase-like protein (At4g27570), partial
(25%)
Length = 577
Score = 95.5 bits (236), Expect = 5e-20
Identities = 59/111 (53%), Positives = 67/111 (60%)
Frame = +3
Query: 366 FSGVLCESLWAWFDVGVFDI*QADGGGPAHG*PSDEFQVGC**AGSGSCGGKRGR*WVGF 425
F GVLCE LW W VGVFD *QAD G G PS E QV C* SG GG+RG+ WVG
Sbjct: 6 FCGVLCEPLWVWVYVGVFDE*QADSVGSTVGGPSFEHQVAC*GTWSGC*GGERGK-WVGL 182
Query: 426 QRGIE*SH*VGDGGR**VGCQTEKQPGHVEGKGLVSKGIKWLHR*VCSRAS 476
QR E SH*V DG R * C +E++ VE S +WLH *VC ++S
Sbjct: 183 QREFEQSH*VSDGWRQ*GWC*SEEESHGVEENWGQS*FNEWLHG*VCPKSS 335
>BG652580 similar to GP|22759895|dbj rhamnosyl transferase {Nierembergia sp.
NB17}, partial (4%)
Length = 404
Score = 94.0 bits (232), Expect = 1e-19
Identities = 53/133 (39%), Positives = 79/133 (58%), Gaps = 2/133 (1%)
Frame = +2
Query: 124 FDVAY-WVPEAARKLGIKTICYHVVSAMSMAMSFVPAKIIP-KDRPLTVEDMSQPPPGYP 181
FD A W+P+ A +LGIK++ + SA+S + VP+++ + R +T ED+ +PPPGYP
Sbjct: 2 FDFAQNWLPKLASELGIKSVRFASFSAISDSYITVPSRLADIEGRNITFEDLKKPPPGYP 181
Query: 182 SSSNSKLVLRRYEAEQQMFASIPFGEGGVTLYDRTTTGLRECDAIAIKTTTEVEGCFCDY 241
+SN + L+ +EA MF FGE T Y+R G +C I ++ E+E + DY
Sbjct: 182 QNSN--ISLKAFEAMDLMFLFKRFGEKNFTGYERVLQGFSDCSLIVFRSCKEIEESYLDY 355
Query: 242 IAIQYHKNVLLTG 254
I Q+ K VLLTG
Sbjct: 356 IEKQFGKLVLLTG 394
>BG237605 weakly similar to GP|8809576|dbj| flavonol
3-O-glucosyltransferase-like {Arabidopsis thaliana},
partial (9%)
Length = 442
Score = 88.6 bits (218), Expect = 6e-18
Identities = 48/131 (36%), Positives = 75/131 (56%), Gaps = 1/131 (0%)
Frame = +3
Query: 24 FAMGHMTPYVHLANELAKRGHKITFLLPNKARLA-LHHLNRYPNLITFHIITVPNVDGLP 82
+ MGH + A+RG I +L+ L+ L L YP ++TF I VP V+GL
Sbjct: 48 YGMGHFPRKTKRFLQRAQRG-PIVYLIGLAGSLSRLQRLLLYPIIVTFVPIKVPYVNGLA 224
Query: 83 LGTETASEIPLSLNHLLVVAMDKTRNQVEQVMKSKKPHFVLFDVAYWVPEAARKLGIKTI 142
ET S++P SL+ L+ AMD+T +E +++ KP V FD +W+P R+LGIK++
Sbjct: 225 HDAETTSDVPFSLSPLIAEAMDRTEKDIEILLRELKPQIVFFDFQHWLPNLTRRLGIKSV 404
Query: 143 CYHVVSAMSMA 153
Y +++ +S A
Sbjct: 405 MYVIINPLSTA 437
>TC234407 weakly similar to GB|AAO63454.1|28951061|BT005390 At5g49690
{Arabidopsis thaliana;} , partial (10%)
Length = 407
Score = 75.1 bits (183), Expect = 7e-14
Identities = 43/132 (32%), Positives = 74/132 (55%), Gaps = 8/132 (6%)
Frame = +2
Query: 16 LHIAMFPWFAMGHMTPYVHLANELAKRGHKITFLLPNKARLALHHLNRYPNLITFHI--- 72
LHIAM PW A+GH+ PY+ L+ LA++GH +TF+ K + + + P + I
Sbjct: 11 LHIAMLPWLAVGHVNPYLELSKILAQKGHFVTFISTPK---NIDGMPKIPETLQPSIKLV 181
Query: 73 -ITVPNVD---GLPLGTETASEIPLSLNHLLVVAMDKTRNQVEQVMKSKKPHFVLFDVA- 127
+ +P+ D LP E+ +IP + ++ L +A + + V +++K+ KP +V +D A
Sbjct: 182 RLPLPHTDHHHHLPEDAESTMDIPSNKSYYLKLAYEALQGPVSELLKTSKPDWVFYDFAT 361
Query: 128 YWVPEAARKLGI 139
W+P A+ L I
Sbjct: 362 EWLPPIAKSLNI 397
>BI426477 weakly similar to GP|10177261|dbj flavonol
3-O-glucosyltransferase-like protein {Arabidopsis
thaliana}, partial (17%)
Length = 406
Score = 74.7 bits (182), Expect = 9e-14
Identities = 43/105 (40%), Positives = 58/105 (54%)
Frame = +2
Query: 213 YDRTTTGLRECDAIAIKTTTEVEGCFCDYIAIQYHKNVLLTGPVLSEEAEGEELEERWAK 272
YDR GL DAI K E+EG + DY+ Q+ K+VLL+GP++ E LE +W
Sbjct: 8 YDRLYGGLSLSDAIGFKGCREIEGPYVDYLEEQFGKSVLLSGPII-PEPPNTVLEGKWGS 184
Query: 273 WLDGLGLGLGLGLGLGPVVYCAFGSQVILGKAQFQEILLGLEISG 317
WL+ V++CA GS+ L QFQE LLGLE++G
Sbjct: 185 WLE--------RFKPDSVIFCALGSEWKLPHDQFQEFLLGLELTG 295
>TC212551 similar to UP|Q9LSM0 (Q9LSM0) Anthocyanidin-3-glucoside
rhamnosyltransferase, partial (9%)
Length = 1037
Score = 46.2 bits (108), Expect(2) = 1e-11
Identities = 30/99 (30%), Positives = 48/99 (48%), Gaps = 7/99 (7%)
Frame = +3
Query: 51 PNKARLALHHLNRYPNL------ITFHIITVPNVDGLPLGTETASEIPLSLNHLLVVAMD 104
P + R+ ++ R P I F + +P V LP E ++IP + L A D
Sbjct: 162 PREFRIHPRNIERLPKPSLNTLDINFVNLPLPKVQNLPENAEANTDIPYDVFEHLKEAYD 341
Query: 105 KTRNQVEQVMKSKKPHFVLFDVA-YWVPEAARKLGIKTI 142
+ +++ ++S KP V +D A +WV A KLGIK +
Sbjct: 342 VLQEPLKRFLESSKPD*VFYDFAPFWVGSMASKLGIKAL 458
Score = 41.2 bits (95), Expect(2) = 1e-11
Identities = 15/30 (50%), Positives = 22/30 (73%)
Frame = +1
Query: 20 MFPWFAMGHMTPYVHLANELAKRGHKITFL 49
MFPW A GHM P + LA +A++GH ++F+
Sbjct: 88 MFPWLAFGHMIPNLELAKLIAQKGHHVSFV 177
>TC229865 weakly similar to UP|P93789 (P93789) UDP-glucose
glucosyltransferase , partial (15%)
Length = 647
Score = 56.2 bits (134), Expect = 3e-08
Identities = 48/171 (28%), Positives = 77/171 (44%), Gaps = 5/171 (2%)
Frame = +1
Query: 11 SSNSKLHIAMFPWFAMGHMTPYVHLANELAKRGHKITFL-LPNKARLALHHLNRYPNLIT 69
+ N +LH+ FP+ A GH+ P + LA A RG K T + P L + + I
Sbjct: 91 NENRELHVLFFPFPANGHIIPSIDLARVFASRGIKTTVVTTPLNVPLISRTIGKAN--IK 264
Query: 70 FHIITVPNVD--GLPLGTETA-SEIPLSLNHLLVVAMDKTRNQVEQVMKSKKPHFVLFDV 126
I P+ + GLP G E + S + L + A R+ +E +M+ + P V+ D+
Sbjct: 265 IKTIKFPSHEETGLPEGCENSDSALSSDLIMTFLKATVLLRDPLENLMQQEHPDCVIADM 444
Query: 127 AY-WVPEAARKLGIKTICYHVVSAMSMAMSFVPAKIIPKDRPLTVEDMSQP 176
Y W ++A K GI + +H + +S P+D V S+P
Sbjct: 445 FYPWATDSAAKFGIPRVVFHGMGFFPTCVSACVRTYKPQD---NVSSWSEP 588
>TC205339 weakly similar to UP|Q6QDB6 (Q6QDB6) UDP-glucose
glucosyltransferase, partial (55%)
Length = 1537
Score = 54.3 bits (129), Expect = 1e-07
Identities = 81/337 (24%), Positives = 131/337 (38%), Gaps = 32/337 (9%)
Frame = +2
Query: 13 NSKLHIAMFPWFAMGHMTPYVHLANELAKRGHKITFLLPNKARLALHHLNRYPNLITFHI 72
+ +LHI +FP+ GH+ P +A RG + T + + I
Sbjct: 20 DGELHIMLFPFPGQGHLIPMSDMARAFNGRGVRTTIVTTPLNVATIRGTIGKETETDIEI 199
Query: 73 ITV--PNVD-GLPLGTETASEIP---LSLNHLLVVAMDKTRNQVEQVMKSKKPHFVLFDV 126
+TV P+ + GLP G E IP L L L + M +E ++ +PH ++
Sbjct: 200 LTVKFPSAEAGLPEGCENTESIPSPDLVLTFLKAIRM--LEAPLEHLLLQHRPHCLIASA 373
Query: 127 AY-WVPEAARKLGIKTICYHVVSAMSMAMS-----FVPAKIIPKDR-PLTVEDMSQPPPG 179
+ W +A KL I + +H ++ S + P K + D P + + PG
Sbjct: 374 FFPWASHSATKLKIPRLVFHGTGVFALCASECVRLYQPHKNVSSDTDPFIIPHL----PG 541
Query: 180 YPSSSNSKLVLRRYEAEQQMFASIPFGEGGVTLYDRTTTGLRECD----AIAIKTTTEVE 235
++L+L Y GE G+T R ++E + + + + E+E
Sbjct: 542 --DIQMTRLLLPDYAKTD------GDGETGLT---RVLQEIKESELASYGMIVNSFYELE 688
Query: 236 GCFCDYIAIQYHKNVL--------LTGPV-LSEEAEGEELEERWA------KWLDGLGLG 280
+ DY Y K +L GP+ L + +G+ ++ KWLD
Sbjct: 689 QVYADY----YDKQLLQVQGRRAWYIGPLSLCNQDKGKRGKQASVDQGDILKWLD----- 841
Query: 281 LGLGLGLGPVVYCAFGSQVILGKAQFQEILLGLEISG 317
VVY FGS + Q +EI GLE SG
Sbjct: 842 ---SKKANSVVYVCFGSIANFSETQLREIARGLEDSG 943
>TC231162 weakly similar to UP|Q9FUJ6 (Q9FUJ6) UDP-glucosyltransferase HRA25,
partial (35%)
Length = 912
Score = 53.9 bits (128), Expect = 2e-07
Identities = 75/324 (23%), Positives = 140/324 (43%), Gaps = 24/324 (7%)
Frame = +2
Query: 17 HIAMFPWFAMGHMTPYVHLANELAKRGHKITFLLPN-KARLALHHLNRYPNLITFHIITV 75
H+ + P+ A GH+ P + L+ +LA+ G K+TF+ + + L N + + +I++
Sbjct: 5 HVLVLPFPAQGHVNPLMLLSKKLAEHGFKVTFVNTDFNHKRVLSATNEEGSAV--RLISI 178
Query: 76 PNVDGLPLGTETASEIPL---SLNHLLVVAMDKTRNQVEQV-MKSKKPHFVLFDV-AYWV 130
P DGL + + + L SL+ + A++K ++ + S+K ++ DV W
Sbjct: 179 P--DGLGPEDDRNNVVNLCSESLSSTMTSALEKVIKDIDALDSASEKITGIVADVNMAWA 352
Query: 131 PEAARKLGIKTICYHVVSAMSMAMSFVPAKIIPKDRPLTVEDMSQPPPGYPSSSNSKLVL 190
E KLGIK + SA + ++ ++ P ++D G+P ++
Sbjct: 353 LELTDKLGIKGAVFCPASAAVL--------VLGENIPNLIQDGIINTEGFP------IIK 490
Query: 191 RRYEAEQQM----FASIPFGEGG-----VTLYDRTTTGLRE---CDAIAIKTTTEVEGCF 238
+++ +M A IP+ G +Y+ + +R D TT+++E
Sbjct: 491 GKFQLSPEMPIMDTADIPWCSLGDPTMHKVIYNHASKIIRYSHLTDWWLGNTTSDLEPG- 667
Query: 239 CDYIAIQYHKNVLLTGPVLSEEAEGEELEERWAK------WLDGLGLGLGLGLGLGPVVY 292
AI +L GP++ + L + W + WLD V+Y
Sbjct: 668 ----AISLSPKILPIGPLIGSGNDIRSLGQFWEEDVSCLTWLD--------QQPPCSVIY 811
Query: 293 CAFGSQVILGKAQFQEILLGLEIS 316
AFGS I Q +E+ LGL+++
Sbjct: 812 VAFGSSTIFDPHQLKELALGLDLT 883
>TC208544 weakly similar to PIR|D96766|D96766 protein glucosyltransferase
F2P9.25 [imported] - Arabidopsis thaliana {Arabidopsis
thaliana;} , partial (16%)
Length = 937
Score = 52.0 bits (123), Expect = 6e-07
Identities = 76/331 (22%), Positives = 124/331 (36%), Gaps = 21/331 (6%)
Frame = +1
Query: 8 NTSSSNSKLHIAMFPWFAMGHMTPYVHLANELAKRGHKITFLLP--NKARLALHHLNRYP 65
+ + S + H+ +P+ GH+ P + L RG +T L+ N+A L Y
Sbjct: 4 HANMSTATTHVLAYPFPTSGHVIPLLDFTKTLVSRGVHVTVLVTPYNEALLP----KNYS 171
Query: 66 NLITFHIITVPNVDGLPLGTETASEIPLSLNHLLVVAMDKTRNQVEQVMKSKKPHFVLFD 125
L+ ++ P P S + +H + MD + Q P ++ D
Sbjct: 172 PLLQTLLLPEPQFPN-PKQNRLVSMVTFMRHHHYPIIMDWAQAQ------PIPPAAIISD 330
Query: 126 VAY-WVPEAARKLGIKTICYHVVSAMSMAMSFVPAKIIPKDRPLTVEDMSQPPPG---YP 181
W AR L + + + A ++++S+ + P++ D + P G +P
Sbjct: 331 FFLGWTHLLARDLHVPRVVFSPSGAFALSVSYSLWRDAPQN------DNPEDPNGVVSFP 492
Query: 182 SSSNSKLVLRRYEAEQQMFASIPFGEGGVTLYDRTTTGLRECDA--IAIKTTTEVEGCFC 239
+ NS Y Q GG L D+ + I T TE+E +
Sbjct: 493 NLPNSPF----YPWWQITHLFHDTERGGPEWKFHRENMLLNIDSWGVVINTFTELEQVYL 660
Query: 240 DYIAIQY-HKNVLLTGPVLS----------EEAEGEELEERW--AKWLDGLGLGLGLGLG 286
+++ + H+ V GPVL EE G R +WLD G
Sbjct: 661 NHLKKELGHERVFAVGPVLPIQTGSISTKPEERGGNSTVSRHDIMEWLDARDKG------ 822
Query: 287 LGPVVYCAFGSQVILGKAQFQEILLGLEISG 317
VVY FGS+ L +Q + + LEISG
Sbjct: 823 --SVVYVCFGSRTFLTSSQMEVLTRALEISG 909
>TC220510 similar to UP|Q7XZD0 (Q7XZD0) Isoflavonoid glucosyltransferase,
partial (62%)
Length = 925
Score = 48.1 bits (113), Expect = 9e-06
Identities = 35/141 (24%), Positives = 69/141 (48%), Gaps = 4/141 (2%)
Frame = +3
Query: 16 LHIAMFPWFAMGHMTPYVHLANELAKRGHKITFL-LPNKARLALHHLNRYPNLITFHIIT 74
L + + A GHM P +A + RGH +T + P+ A++ L +P L+ H +
Sbjct: 9 LKLYFIHFLAEGHMIPLCDMATLFSTRGHHVTIITTPSNAQILRKSLPSHP-LLRLHTVQ 185
Query: 75 VPNVD-GLPLGTETA-SEIPLSLNHLLVVAMDKTRNQVEQVMKSKKPHFVLFDVAY-WVP 131
P+ + GLP G E ++ L + A + +E +++ + P ++ D + WV
Sbjct: 186 FPSHEVGLPDGIENIYADSDLDSLRKVCSATAMLQPPIEDLVEQQPPDCIVADYLFPWVD 365
Query: 132 EAARKLGIKTICYHVVSAMSM 152
+ A+KL I + ++ +S ++
Sbjct: 366 DLAKKLHIPRLAFNGLSLFTI 428
>TC203711 similar to UP|Q8S9A4 (Q8S9A4) Glucosyltransferase-5, partial (57%)
Length = 1322
Score = 41.6 bits (96), Expect(2) = 1e-05
Identities = 36/102 (35%), Positives = 50/102 (48%)
Frame = +2
Query: 322 GGSEGSGRLRNR**GVAGGV*REGEGKGNGFERVGPTVVDLEAWFSGVLCESLWAWFDVG 381
GG SGR+ VA V E +G+GNG E +GPT D E+ G + +SL VG
Sbjct: 641 GGGTESGRV------VARRVFGEDQGEGNGGEGLGPTGGDSESRLGGWVRDSLRLELGVG 802
Query: 382 VFDI*QADGGGPAHG*PSDEFQVGC**AGSGSCGGKRGR*WV 423
+*+ GG A +++ Q G G GG+R + WV
Sbjct: 803 -SGV*RGADGGVASLRGAEDEQDGYGEGNEGGFGGEREQGWV 925
Score = 25.4 bits (54), Expect(2) = 1e-05
Identities = 13/27 (48%), Positives = 16/27 (59%)
Frame = +3
Query: 290 VVYCAFGSQVILGKAQFQEILLGLEIS 316
VV FGS +AQ +EI +GLE S
Sbjct: 507 VVLLCFGSMGRFSRAQLKEIAIGLEKS 587
>TC210384 weakly similar to UP|Q8VZF9 (Q8VZF9) AT5g12890/T24H18_60, partial
(11%)
Length = 860
Score = 44.7 bits (104), Expect = 9e-05
Identities = 35/154 (22%), Positives = 68/154 (43%), Gaps = 10/154 (6%)
Frame = +2
Query: 10 SSSNSKLHIAMFPWFAMGHMTPYVHLANELAKRGHKITFLLPNKARLALHHLNRYPNLIT 69
+ + K +FP+ A GH+ P++ LA EL +R K + + N + + P T
Sbjct: 140 AETEGKQEAVLFPFMAQGHIIPFLALALELEQR-KKYSITILNTSLNIKKLRSSIPPDST 316
Query: 70 FHIITV---PNVDGLPLGTETASEIPLSLNHLLVVA----MDKTRNQVEQVMKSKKPH-- 120
++ + P+ GLP TE IP L L+ A + ++ ++ + H
Sbjct: 317 ISLVEIPFTPSDHGLPPNTENTDSIPYHLVIRLIQASTTLQPAFKTLIQNILFQNQKHQL 496
Query: 121 FVLFDVAY-WVPEAARKLGIKTICYHVVSAMSMA 153
++ D+ + W A++LG+ + + S +A
Sbjct: 497 LIISDIFFGWTATVAKELGVFHVVFSGTSGFGLA 598
>BI973419
Length = 423
Score = 44.3 bits (103), Expect = 1e-04
Identities = 43/147 (29%), Positives = 65/147 (43%), Gaps = 13/147 (8%)
Frame = +2
Query: 7 KNTSSSNSKLHIAMFPWFAMGHMTPYVHLANELAKRGHKITFLLP--NKARL--ALHHLN 62
K TS+ S H + P+ +GHM P + + LA G KITFL+ N+ R+ + HL
Sbjct: 2 KFTSTEMSYPHFLVMPYPILGHMNPLLQFSQVLANHGCKITFLITEFNQKRMKSEIDHLG 181
Query: 63 RYPNLITFHIITVPNVDGLPLGTETASE--IPLSLN-------HLLVVAMDKTRNQVEQV 113
N +T DGL + + + + LSL H L+ ++ N ++
Sbjct: 182 AQINFVTL-------PDGLDPEDDRSDQPKVILSLRNTMPTKLHRLIQDINNNNNALDG- 337
Query: 114 MKSKKPHFVLFDVAYWVPEAARKLGIK 140
+K V+ W E A KLGIK
Sbjct: 338 DNNKITCLVVSKNIGWALEVAHKLGIK 418
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.334 0.148 0.485
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 22,191,890
Number of Sequences: 63676
Number of extensions: 326177
Number of successful extensions: 3920
Number of sequences better than 10.0: 118
Number of HSP's better than 10.0 without gapping: 3147
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3606
length of query: 484
length of database: 12,639,632
effective HSP length: 101
effective length of query: 383
effective length of database: 6,208,356
effective search space: 2377800348
effective search space used: 2377800348
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.5 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0282.12


