

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0258c.2
(986 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
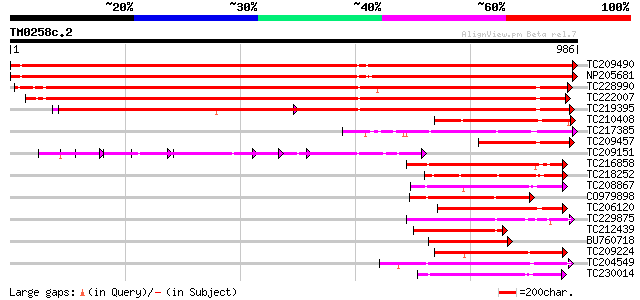
Score E
Sequences producing significant alignments: (bits) Value
TC209490 UP|Q8GSN9 (Q8GSN9) LRR receptor-like kinase (Fragment),... 1538 0.0
NP205681 receptor protein kinase-like protein 1509 0.0
TC228990 UP|Q9LKZ6 (Q9LKZ6) Receptor-like protein kinase 1, comp... 959 0.0
TC222007 UP|Q9LKZ5 (Q9LKZ5) Receptor-like protein kinase 2, comp... 951 0.0
TC219395 UP|Q9LKZ4 (Q9LKZ4) Receptor-like protein kinase 3, part... 926 0.0
TC210408 similar to UP|Q9FEU2 (Q9FEU2) Receptor protein kinase, ... 244 1e-64
TC217385 similar to UP|Q6ZGC7 (Q6ZGC7) Receptor protein kinase P... 237 2e-62
TC209457 homologue to UP|Q9LKZ6 (Q9LKZ6) Receptor-like protein k... 227 2e-59
TC209151 similar to UP|Q9FGN6 (Q9FGN6) Receptor protein kinase-l... 224 1e-58
TC216858 homologue to UP|Q76FZ8 (Q76FZ8) Brassinosteroid recepto... 218 1e-56
TC218252 weakly similar to UP|Q03407 (Q03407) Ydr490cp, partial ... 206 5e-53
TC208867 similar to UP|Q9LXT2 (Q9LXT2) Serine/threonine-specific... 205 9e-53
CO979898 204 1e-52
TC206120 similar to UP|Q941F6 (Q941F6) Leucine-rich repeat recep... 199 5e-51
TC229875 weakly similar to UP|Q75WU3 (Q75WU3) Leucine-rich repea... 198 9e-51
TC212439 similar to UP|Q9LVP0 (Q9LVP0) Receptor-like protein kin... 194 2e-49
BU760718 similar to GP|3641252|gb| leucine-rich receptor-like pr... 190 3e-48
TC209224 similar to UP|BRI1_LYCPE (Q8L899) Systemin receptor SR1... 189 4e-48
TC204549 similar to UP|Q9M0X5 (Q9M0X5) Receptor protein kinase-l... 188 1e-47
TC230014 similar to GB|AAP21294.1|30102752|BT006486 At5g49760 {A... 187 2e-47
>TC209490 UP|Q8GSN9 (Q8GSN9) LRR receptor-like kinase (Fragment), complete
Length = 3298
Score = 1538 bits (3983), Expect = 0.0
Identities = 773/991 (78%), Positives = 868/991 (87%), Gaps = 5/991 (0%)
Frame = +1
Query: 1 MRIRVSYLLVLCFTLIWFRWTVVYSSFSDLDALLKLKESMKGAKAKHHALEDWKFSTSLS 60
MR V Y L+L IW R SSF+D+++LLKLK+SMKG KAK AL DWKF SLS
Sbjct: 91 MRSCVCYTLLLFIFFIWLR-VATCSSFTDMESLLKLKDSMKGDKAKDDALHDWKFFPSLS 267
Query: 61 AHCSFSGVTCDQNLRVVALNVTLVPLFGHLPPEIGLLEKLENLTISMNNLTDQLPSDLAS 120
AHC FSGV CD+ LRVVA+NV+ VPLFGHLPPEIG L+KLENLT+S NNLT LP +LA+
Sbjct: 268 AHCFFSGVKCDRELRVVAINVSFVPLFGHLPPEIGQLDKLENLTVSQNNLTGVLPKELAA 447
Query: 121 LTSLKVLNISHNLFSGQFPGNITVGMTELEALDAYDNSFSGPLPEEIVKLEKLKYLHLAG 180
LTSLK LNISHN+FSG FPG I + MT+LE LD YDN+F+GPLP E+VKLEKLKYL L G
Sbjct: 448 LTSLKHLNISHNVFSGHFPGQIILPMTKLEVLDVYDNNFTGPLPVELVKLEKLKYLKLDG 627
Query: 181 NYFSGTIPESYSEFQSLEFLGLNANSLTGRVPESLAKLKTLKELHLGYSNAYEGGIPPAF 240
NYFSG+IPESYSEF+SLEFL L+ NSL+G++P+SL+KLKTL+ L LGY+NAYEGGIPP F
Sbjct: 628 NYFSGSIPESYSEFKSLEFLSLSTNSLSGKIPKSLSKLKTLRYLKLGYNNAYEGGIPPEF 807
Query: 241 GSMENLRLLEMANCNLTGEIPPSLGNLTKLHSLFVQMNNLTGTIPPELSSMMSLMSLDLS 300
GSM++LR L++++CNL+GEIPPSL NLT L +LF+Q+NNLTGTIP ELS+M+SLMSLDLS
Sbjct: 808 GSMKSLRYLDLSSCNLSGEIPPSLANLTNLDTLFLQINNLTGTIPSELSAMVSLMSLDLS 987
Query: 301 INDLTGEIPESFSKLKNLTLMNFFQNKFRGSLPSFIGDLPNLETLQVWENNFSFVLPHNL 360
INDLTGEIP SFS+L+NLTLMNFFQN RGS+PSF+G+LPNLETLQ+W+NNFSFVLP NL
Sbjct: 988 INDLTGEIPMSFSQLRNLTLMNFFQNNLRGSVPSFVGELPNLETLQLWDNNFSFVLPPNL 1167
Query: 361 GGNGRFLYFDVTKNHLTGLIPPDLCKSGRLKTFIITDNFFRGPIPKGIGECRSLTKIRVA 420
G NG+ +FDV KNH TGLIP DLCKSGRL+T +ITDNFFRGPIP IG C+SLTKIR +
Sbjct: 1168 GQNGKLKFFDVIKNHFTGLIPRDLCKSGRLQTIMITDNFFRGPIPNEIGNCKSLTKIRAS 1347
Query: 421 NNFLDGPVPPGVFQLPSVTITELSNNRLNGELPSVISGESLGTLTLSNNLFTGKIPAAMK 480
NN+L+G VP G+F+LPSVTI EL+NNR NGELP ISGESLG LTLSNNLF+GKIP A+K
Sbjct: 1348 NNYLNGVVPSGIFKLPSVTIIELANNRFNGELPPEISGESLGILTLSNNLFSGKIPPALK 1527
Query: 481 NLRALQSLSLDANEFIGEIPGGVFEIPMLTKVNISGNNLTGPIPTTITHRASLTAVDLSR 540
NLRALQ+LSLDANEF+GEIPG VF++PMLT VNISGNNLTGPIPTT+T SLTAVDLSR
Sbjct: 1528 NLRALQTLSLDANEFVGEIPGEVFDLPMLTVVNISGNNLTGPIPTTLTRCVSLTAVDLSR 1707
Query: 541 NNLAGEVPKGMKNLMDLSILNLSRNEISGPVPDEIRFMTSLTTLDLSSNNFTGTVPTGGQ 600
N L G++PKG+KNL DLSI N+S N+ISGPVP+EIRFM SLTTLDLS+NNF G VPTGGQ
Sbjct: 1708 NMLEGKIPKGIKNLTDLSIFNVSINQISGPVPEEIRFMLSLTTLDLSNNNFIGKVPTGGQ 1887
Query: 601 FLVFNYDKTFAGNPNLCFPHRASCP-SVLY--DSLRKTRA--KTARVRAIVIGIALATAV 655
F VF+ +K+FAGNPNLC H SCP S LY D+L+K R R IVI IAL TA
Sbjct: 1888 FAVFS-EKSFAGNPNLCTSH--SCPNSSLYPDDALKKRRGPWSLKSTRVIVIVIALGTAA 2058
Query: 656 LLVAVTVHVVRKRRLHRAQAWKLTAFQRLEIKAEDVVECLKEENIIGKGGAGIVYRGSMP 715
LLVAVTV+++R+R+++ A+ WKLTAFQRL KAEDVVECLKEENIIGKGGAGIVYRGSMP
Sbjct: 2059 LLVAVTVYMMRRRKMNLAKTWKLTAFQRLNFKAEDVVECLKEENIIGKGGAGIVYRGSMP 2238
Query: 716 NGTDVAIKRLVGQGSGRNDYGFRAEIETLGKIRHRNIMRLLGYVSNKDTNLLLYEYMPNG 775
NGTDVAIKRLVG GSGRNDYGF+AEIETLGKIRHRNIMRLLGYVSNK+TNLLLYEYMPNG
Sbjct: 2239 NGTDVAIKRLVGAGSGRNDYGFKAEIETLGKIRHRNIMRLLGYVSNKETNLLLYEYMPNG 2418
Query: 776 SLGEWLHGAKGGHLRWEMRYKIAVEAARGLCYMHHDCSPLIIHRDVKSNNILLDADFEAH 835
SLGEWLHGAKGGHL+WEMRYKIAVEAA+GLCY+HHDCSPLIIHRDVKSNNILLD D EAH
Sbjct: 2419 SLGEWLHGAKGGHLKWEMRYKIAVEAAKGLCYLHHDCSPLIIHRDVKSNNILLDGDLEAH 2598
Query: 836 VADFGLAKFLYDPGASQSMSSIAGSYGYIAPEYAYTLKVDEKSDVYSFGVVLLELIIGRK 895
VADFGLAKFLYDPGASQSMSSIAGSYGYIAPEYAYTLKVDEKSDVYSFGVVLLELIIGRK
Sbjct: 2599 VADFGLAKFLYDPGASQSMSSIAGSYGYIAPEYAYTLKVDEKSDVYSFGVVLLELIIGRK 2778
Query: 896 PVGEFGDGVDIVGWVNKTMSELSQPSDTALVLAVVDPRLSGYPLTSVIHMFNIAMMCVKE 955
PVGEFGDGVDIVGWVNKT EL+QPSD ALVLAVVDPRLSGYPLTSVI+MFNIAMMCVKE
Sbjct: 2779 PVGEFGDGVDIVGWVNKTRLELAQPSDAALVLAVVDPRLSGYPLTSVIYMFNIAMMCVKE 2958
Query: 956 MGPARPTMREVVHMLTNPPQSNTSTQDLINL 986
MGPARPTMREVVHML+ PP S T T +LINL
Sbjct: 2959 MGPARPTMREVVHMLSEPPHSATHTHNLINL 3051
>NP205681 receptor protein kinase-like protein
Length = 2946
Score = 1509 bits (3906), Expect = 0.0
Identities = 761/988 (77%), Positives = 848/988 (85%), Gaps = 2/988 (0%)
Frame = +1
Query: 1 MRIRVSYLLVLCFTLIWFRWTVVYSSFSDLDALLKLKESMKGAKAKHHALEDWKFSTSLS 60
MR V Y L+L IW SSFSD+DALLKLKESMKG +AK AL DWKFSTSLS
Sbjct: 1 MRSCVCYTLLLFVFFIWLH-VATCSSFSDMDALLKLKESMKGDRAKDDALHDWKFSTSLS 177
Query: 61 AHCSFSGVTCDQNLRVVALNVTLVPLFGHLPPEIGLLEKLENLTISMNNLTDQLPSDLAS 120
AHC FSGV+CDQ LRVVA+NV+ VPLFGH+PPEIG L+KLENLTIS NNLT +LP +LA+
Sbjct: 178 AHCFFSGVSCDQELRVVAINVSFVPLFGHVPPEIGELDKLENLTISQNNLTGELPKELAA 357
Query: 121 LTSLKVLNISHNLFSGQFPGNITVGMTELEALDAYDNSFSGPLPEEIVKLEKLKYLHLAG 180
LTSLK LNISHN+FSG FPG I + MTELE LD YDN+F+G LPEE VKLEKLKYL L G
Sbjct: 358 LTSLKHLNISHNVFSGYFPGKIILPMTELEVLDVYDNNFTGSLPEEFVKLEKLKYLKLDG 537
Query: 181 NYFSGTIPESYSEFQSLEFLGLNANSLTGRVPESLAKLKTLKELHLGYSNAYEGGIPPAF 240
NYFSG+IPESYSEF+SLEFL L+ NSL+G +P+SL+KLKTL+ L LGY+NAYEGGIPP F
Sbjct: 538 NYFSGSIPESYSEFKSLEFLSLSTNSLSGNIPKSLSKLKTLRILKLGYNNAYEGGIPPEF 717
Query: 241 GSMENLRLLEMANCNLTGEIPPSLGNLTKLHSLFVQMNNLTGTIPPELSSMMSLMSLDLS 300
G+ME+L+ L++++CNL+GEIPPSL N+ L +LF+QMNNLTGTIP ELS M+SLMSLDLS
Sbjct: 718 GTMESLKYLDLSSCNLSGEIPPSLANMRNLDTLFLQMNNLTGTIPSELSDMVSLMSLDLS 897
Query: 301 INDLTGEIPESFSKLKNLTLMNFFQNKFRGSLPSFIGDLPNLETLQVWENNFSFVLPHNL 360
N LTGEIP FS+LKNLTLMNFF N RGS+PSF+G+LPNLETLQ+WENNFS LP NL
Sbjct: 898 FNGLTGEIPTRFSQLKNLTLMNFFHNNLRGSVPSFVGELPNLETLQLWENNFSSELPQNL 1077
Query: 361 GGNGRFLYFDVTKNHLTGLIPPDLCKSGRLKTFIITDNFFRGPIPKGIGECRSLTKIRVA 420
G NG+F +FDVTKNH +GLIP DLCKSGRL+TF+ITDNFF GPIP I C+SLTKIR +
Sbjct: 1078GQNGKFKFFDVTKNHFSGLIPRDLCKSGRLQTFLITDNFFHGPIPNEIANCKSLTKIRAS 1257
Query: 421 NNFLDGPVPPGVFQLPSVTITELSNNRLNGELPSVISGESLGTLTLSNNLFTGKIPAAMK 480
NN+L+G VP G+F+LPSVTI EL+NNR NGELP ISG+SLG LTLSNNLFTGKIP A+K
Sbjct: 1258NNYLNGAVPSGIFKLPSVTIIELANNRFNGELPPEISGDSLGILTLSNNLFTGKIPPALK 1437
Query: 481 NLRALQSLSLDANEFIGEIPGGVFEIPMLTKVNISGNNLTGPIPTTITHRASLTAVDLSR 540
NLRALQ+LSLD NEF+GEIPG VF++PMLT VNISGNNLTGPIPTT T SL AVDLSR
Sbjct: 1438NLRALQTLSLDTNEFLGEIPGEVFDLPMLTVVNISGNNLTGPIPTTFTRCVSLAAVDLSR 1617
Query: 541 NNLAGEVPKGMKNLMDLSILNLSRNEISGPVPDEIRFMTSLTTLDLSSNNFTGTVPTGGQ 600
N L GE+PKGMKNL DLSI N+S N+ISG VPDEIRFM SLTTLDLS NNF G VPTGGQ
Sbjct: 1618NMLDGEIPKGMKNLTDLSIFNVSINQISGSVPDEIRFMLSLTTLDLSYNNFIGKVPTGGQ 1797
Query: 601 FLVFNYDKTFAGNPNLCFPHRASCPSVLYDSLRKTRA--KTARVRAIVIGIALATAVLLV 658
FLVF+ DK+FAGNPNLC H SCP+ SL+K R R IV+ IALATA +LV
Sbjct: 1798FLVFS-DKSFAGNPNLCSSH--SCPN---SSLKKRRGPWSLKSTRVIVMVIALATAAILV 1959
Query: 659 AVTVHVVRKRRLHRAQAWKLTAFQRLEIKAEDVVECLKEENIIGKGGAGIVYRGSMPNGT 718
A T ++ R+R+L A WKLT FQRL +KAE+VVECLKEENIIGKGGAGIVYRGSM NG+
Sbjct: 1960AGTEYMRRRRKLKLAMTWKLTGFQRLNLKAEEVVECLKEENIIGKGGAGIVYRGSMRNGS 2139
Query: 719 DVAIKRLVGQGSGRNDYGFRAEIETLGKIRHRNIMRLLGYVSNKDTNLLLYEYMPNGSLG 778
DVAIKRLVG GSGRNDYGF+AEIET+GKIRHRNIMRLLGYVSNK+TNLLLYEYMPNGSLG
Sbjct: 2140DVAIKRLVGAGSGRNDYGFKAEIETVGKIRHRNIMRLLGYVSNKETNLLLYEYMPNGSLG 2319
Query: 779 EWLHGAKGGHLRWEMRYKIAVEAARGLCYMHHDCSPLIIHRDVKSNNILLDADFEAHVAD 838
EWLHGAKGGHL+WEMRYKIAVEAA+GLCY+HHDCSPLIIHRDVKSNNILLDA FEAHVAD
Sbjct: 2320EWLHGAKGGHLKWEMRYKIAVEAAKGLCYLHHDCSPLIIHRDVKSNNILLDAHFEAHVAD 2499
Query: 839 FGLAKFLYDPGASQSMSSIAGSYGYIAPEYAYTLKVDEKSDVYSFGVVLLELIIGRKPVG 898
FGLAKFLYD G+SQSMSSIAGSYGYIAPEYAYTLKVDEKSDVYSFGVVLLELIIGRKPVG
Sbjct: 2500FGLAKFLYDLGSSQSMSSIAGSYGYIAPEYAYTLKVDEKSDVYSFGVVLLELIIGRKPVG 2679
Query: 899 EFGDGVDIVGWVNKTMSELSQPSDTALVLAVVDPRLSGYPLTSVIHMFNIAMMCVKEMGP 958
EFGDGVDIVGWVNKT ELSQPSD A+VLAVVDPRLSGYPL SVI+MFNIAMMCVKE+GP
Sbjct: 2680EFGDGVDIVGWVNKTRLELSQPSDAAVVLAVVDPRLSGYPLISVIYMFNIAMMCVKEVGP 2859
Query: 959 ARPTMREVVHMLTNPPQSNTSTQDLINL 986
RPTMREVVHML+NPP T T +LINL
Sbjct: 2860TRPTMREVVHMLSNPPHFTTHTHNLINL 2943
>TC228990 UP|Q9LKZ6 (Q9LKZ6) Receptor-like protein kinase 1, complete
Length = 3346
Score = 959 bits (2480), Expect = 0.0
Identities = 492/978 (50%), Positives = 663/978 (67%), Gaps = 6/978 (0%)
Frame = +1
Query: 8 LLVLCFTLIWFRWTVVYSSFSDLDALLKLKESMKGAKAKHHALEDWKFSTSLSAHCSFSG 67
+LVL F F ++ + S+ ALL K S HAL W ST CS+ G
Sbjct: 106 VLVLFFL---FLHSLQAARISEYRALLSFKASSL-TDDPTHALSSWNSSTPF---CSWFG 264
Query: 68 VTCDQNLRVVALNVTLVPLFGHLPPEIGLLEKLENLTISMNNLTDQLPSDLASLTSLKVL 127
+TCD V +LN+T + L G L ++ L L +L+++ N + +P+ ++L++L+ L
Sbjct: 265 LTCDSRRHVTSLNLTSLSLSGTLSDDLSHLPFLSHLSLADNKFSGPIPASFSALSALRFL 444
Query: 128 NISHNLFSGQFPGNITVGMTELEALDAYDNSFSGPLPEEIVKLEKLKYLHLAGNYFSGTI 187
N+S+N+F+ FP + + LE LD Y+N+ +G LP + + L++LHL GN+FSG I
Sbjct: 445 NLSNNVFNATFPSQLN-RLANLEVLDLYNNNMTGELPLSVAAMPLLRHLHLGGNFFSGQI 621
Query: 188 PESYSEFQSLEFLGLNANSLTGRVPESLAKLKTLKELHLGYSNAYEGGIPPAFGSMENLR 247
P Y +Q L++L L+ N L G + L L +L+EL++GY N Y GGIPP G++ NL
Sbjct: 622 PPEYGTWQHLQYLALSGNELAGTIAPELGNLSSLRELYIGYYNTYSGGIPPEIGNLSNLV 801
Query: 248 LLEMANCNLTGEIPPSLGNLTKLHSLFVQMNNLTGTIPPELSSMMSLMSLDLSINDLTGE 307
L+ A C L+GEIP LG L L +LF+Q+N L+G++ PEL S+ SL S+DLS N L+GE
Sbjct: 802 RLDAAYCGLSGEIPAELGKLQNLDTLFLQVNALSGSLTPELGSLKSLKSMDLSNNMLSGE 981
Query: 308 IPESFSKLKNLTLMNFFQNKFRGSLPSFIGDLPNLETLQVWENNFSFVLPHNLGGNGRFL 367
+P SF++LKNLTL+N F+NK G++P F+G+LP LE LQ+WENNF+ +P NLG NGR
Sbjct: 982 VPASFAELKNLTLLNLFRNKLHGAIPEFVGELPALEVLQLWENNFTGSIPQNLGNNGRLT 1161
Query: 368 YFDVTKNHLTGLIPPDLCKSGRLKTFIITDNFFRGPIPKGIGECRSLTKIRVANNFLDGP 427
D++ N +TG +PP++C RL+T I N+ GPIP +G+C+SL +IR+ NFL+G
Sbjct: 1162 LVDLSSNKITGTLPPNMCYGNRLQTLITLGNYLFGPIPDSLGKCKSLNRIRMGENFLNGS 1341
Query: 428 VPPGVFQLPSVTITELSNNRLNGELPSVIS-GESLGTLTLSNNLFTGKIPAAMKNLRALQ 486
+P G+F LP +T EL +N L G+ P S LG ++LSNN +G +P+ + N ++Q
Sbjct: 1342 IPKGLFGLPKLTQVELQDNLLTGQFPEDGSIATDLGQISLSNNQLSGSLPSTIGNFTSMQ 1521
Query: 487 SLSLDANEFIGEIPGGVFEIPMLTKVNISGNNLTGPIPTTITHRASLTAVDLSRNNLAGE 546
L L+ NEF G IP + + L+K++ S N +GPI I+ LT +DLS N L+GE
Sbjct: 1522 KLLLNGNEFTGRIPPQIGMLQQLSKIDFSHNKFSGPIAPEISKCKLLTFIDLSGNELSGE 1701
Query: 547 VPKGMKNLMDLSILNLSRNEISGPVPDEIRFMTSLTTLDLSSNNFTGTVPTGGQFLVFNY 606
+P + ++ L+ LNLSRN + G +P I M SLT++D S NNF+G VP GQF FNY
Sbjct: 1702 IPNKITSMRILNYLNLSRNHLDGSIPGNIASMQSLTSVDFSYNNFSGLVPGTGQFGYFNY 1881
Query: 607 DKTFAGNPNLCFPHRASCPSVLYDSLRKTRAK----TARVRAIVIGIALATAVLLVAVTV 662
+F GNP LC P+ C + + R+ K ++ +VIG+ + + + VA
Sbjct: 1882 -TSFLGNPELCGPYLGPCKDGVANGPRQPHVKGPFSSSLKLLLVIGLLVCSILFAVAAIF 2058
Query: 663 HVVRKRRLHRAQAWKLTAFQRLEIKAEDVVECLKEENIIGKGGAGIVYRGSMPNGTDVAI 722
++ A+AWKLTAFQRL+ +DV++CLKE+NIIGKGGAGIVY+G+MPNG +VA+
Sbjct: 2059 KARALKKASEARAWKLTAFQRLDFTVDDVLDCLKEDNIIGKGGAGIVYKGAMPNGDNVAV 2238
Query: 723 KRLVGQGSG-RNDYGFRAEIETLGKIRHRNIMRLLGYVSNKDTNLLLYEYMPNGSLGEWL 781
KRL G +D+GF AEI+TLG+IRHR+I+RLLG+ SN +TNLL+YEYMPNGSLGE L
Sbjct: 2239 KRLPAMSRGSSHDHGFNAEIQTLGRIRHRHIVRLLGFCSNHETNLLVYEYMPNGSLGEVL 2418
Query: 782 HGAKGGHLRWEMRYKIAVEAARGLCYMHHDCSPLIIHRDVKSNNILLDADFEAHVADFGL 841
HG KGGHL W+ RYKIAVEAA+GLCY+HHDCSPLI+HRDVKSNNILLD++FEAHVADFGL
Sbjct: 2419 HGKKGGHLHWDTRYKIAVEAAKGLCYLHHDCSPLIVHRDVKSNNILLDSNFEAHVADFGL 2598
Query: 842 AKFLYDPGASQSMSSIAGSYGYIAPEYAYTLKVDEKSDVYSFGVVLLELIIGRKPVGEFG 901
AKFL D GAS+ MS+IAGSYGYIAPEYAYTLKVDEKSDVYSFGVVLLEL+ GRKPVGEFG
Sbjct: 2599 AKFLQDSGASECMSAIAGSYGYIAPEYAYTLKVDEKSDVYSFGVVLLELVTGRKPVGEFG 2778
Query: 902 DGVDIVGWVNKTMSELSQPSDTALVLAVVDPRLSGYPLTSVIHMFNIAMMCVKEMGPARP 961
DGVDIV WV K S+ VL V+D RL PL V+H+F +AM+CV+E RP
Sbjct: 2779 DGVDIVQWVRKMTD-----SNKEGVLKVLDSRLPSVPLHEVMHVFYVAMLCVEEQAVERP 2943
Query: 962 TMREVVHMLTNPPQSNTS 979
TMREVV +LT P+ +S
Sbjct: 2944 TMREVVQILTELPKPPSS 2997
>TC222007 UP|Q9LKZ5 (Q9LKZ5) Receptor-like protein kinase 2, complete
Length = 3192
Score = 951 bits (2459), Expect = 0.0
Identities = 486/953 (50%), Positives = 649/953 (67%), Gaps = 5/953 (0%)
Frame = +3
Query: 28 SDLDALLKLKESMKGAKAKHHALEDWKFSTSLSAHCSFSGVTCDQNLRVVALNVTLVPLF 87
S+ ALL L+ + A L W S +CS+ GVTCD V ALN+T + L
Sbjct: 93 SEYRALLSLRSVITDATPP--VLSSWNASIP---YCSWLGVTCDNRRHVTALNLTGLDLS 257
Query: 88 GHLPPEIGLLEKLENLTISMNNLTDQLPSDLASLTSLKVLNISHNLFSGQFPGNITVGMT 147
G L ++ L L NL+++ N + +P L++L+ L+ LN+S+N+F+ FP + +
Sbjct: 258 GTLSADVAHLPFLSNLSLAANKFSGPIPPSLSALSGLRYLNLSNNVFNETFPSELW-RLQ 434
Query: 148 ELEALDAYDNSFSGPLPEEIVKLEKLKYLHLAGNYFSGTIPESYSEFQSLEFLGLNANSL 207
LE LD Y+N+ +G LP + +++ L++LHL GN+FSG IP Y +Q L++L ++ N L
Sbjct: 435 SLEVLDLYNNNMTGVLPLAVAQMQNLRHLHLGGNFFSGQIPPEYGRWQRLQYLAVSGNEL 614
Query: 208 TGRVPESLAKLKTLKELHLGYSNAYEGGIPPAFGSMENLRLLEMANCNLTGEIPPSLGNL 267
G +P + L +L+EL++GY N Y GGIPP G++ L L++A C L+GEIP +LG L
Sbjct: 615 DGTIPPEIGNLTSLRELYIGYYNTYTGGIPPEIGNLSELVRLDVAYCALSGEIPAALGKL 794
Query: 268 TKLHSLFVQMNNLTGTIPPELSSMMSLMSLDLSINDLTGEIPESFSKLKNLTLMNFFQNK 327
KL +LF+Q+N L+G++ PEL ++ SL S+DLS N L+GEIP SF +LKN+TL+N F+NK
Sbjct: 795 QKLDTLFLQVNALSGSLTPELGNLKSLKSMDLSNNMLSGEIPASFGELKNITLLNLFRNK 974
Query: 328 FRGSLPSFIGDLPNLETLQVWENNFSFVLPHNLGGNGRFLYFDVTKNHLTGLIPPDLCKS 387
G++P FIG+LP LE +Q+WENN + +P LG NGR D++ N LTG +PP LC
Sbjct: 975 LHGAIPEFIGELPALEVVQLWENNLTGSIPEGLGKNGRLNLVDLSSNKLTGTLPPYLCSG 1154
Query: 388 GRLKTFIITDNFFRGPIPKGIGECRSLTKIRVANNFLDGPVPPGVFQLPSVTITELSNNR 447
L+T I NF GPIP+ +G C SLT+IR+ NFL+G +P G+F LP +T EL +N
Sbjct: 1155 NTLQTLITLGNFLFGPIPESLGTCESLTRIRMGENFLNGSIPKGLFGLPKLTQVELQDNY 1334
Query: 448 LNGELPSVIS-GESLGTLTLSNNLFTGKIPAAMKNLRALQSLSLDANEFIGEIPGGVFEI 506
L+GE P V S +LG +TLSNN +G + ++ N ++Q L LD N F G IP + +
Sbjct: 1335 LSGEFPEVGSVAVNLGQITLSNNQLSGALSPSIGNFSSVQKLLLDGNMFTGRIPTQIGRL 1514
Query: 507 PMLTKVNISGNNLTGPIPTTITHRASLTAVDLSRNNLAGEVPKGMKNLMDLSILNLSRNE 566
L+K++ SGN +GPI I+ LT +DLSRN L+G++P + + L+ LNLS+N
Sbjct: 1515 QQLSKIDFSGNKFSGPIAPEISQCKLLTFLDLSRNELSGDIPNEITGMRILNYLNLSKNH 1694
Query: 567 ISGPVPDEIRFMTSLTTLDLSSNNFTGTVPTGGQFLVFNYDKTFAGNPNLCFPHRASCPS 626
+ G +P I M SLT++D S NN +G VP GQF FNY +F GNP+LC P+ +C
Sbjct: 1695 LVGSIPSSISSMQSLTSVDFSYNNLSGLVPGTGQFSYFNY-TSFLGNPDLCGPYLGACKG 1871
Query: 627 VLYDSLRKTRAK---TARVRAIVIGIALATAVLLVAVTVHVVRKRRLHRAQAWKLTAFQR 683
+ + + K ++ +V+G+ L + VA ++ A+AWKLTAFQR
Sbjct: 1872 GVANGAHQPHVKGLSSSLKLLLVVGLLLCSIAFAVAAIFKARSLKKASEARAWKLTAFQR 2051
Query: 684 LEIKAEDVVECLKEENIIGKGGAGIVYRGSMPNGTDVAIKRLVGQGSG-RNDYGFRAEIE 742
L+ +DV+ CLKE+NIIGKGGAGIVY+G+MPNG VA+KRL G +D+GF AEI+
Sbjct: 2052 LDFTVDDVLHCLKEDNIIGKGGAGIVYKGAMPNGDHVAVKRLPAMSRGSSHDHGFNAEIQ 2231
Query: 743 TLGKIRHRNIMRLLGYVSNKDTNLLLYEYMPNGSLGEWLHGAKGGHLRWEMRYKIAVEAA 802
TLG+IRHR+I+RLLG+ SN +TNLL+YEYMPNGSLGE LHG KGGHL W+ RYKIAVEAA
Sbjct: 2232 TLGRIRHRHIVRLLGFCSNHETNLLVYEYMPNGSLGEVLHGKKGGHLHWDTRYKIAVEAA 2411
Query: 803 RGLCYMHHDCSPLIIHRDVKSNNILLDADFEAHVADFGLAKFLYDPGASQSMSSIAGSYG 862
+GLCY+HHDCSPLI+HRDVKSNNILLD++ EAHVADFGLAKFL D G S+ MS+IAGSYG
Sbjct: 2412 KGLCYLHHDCSPLIVHRDVKSNNILLDSNHEAHVADFGLAKFLQDSGTSECMSAIAGSYG 2591
Query: 863 YIAPEYAYTLKVDEKSDVYSFGVVLLELIIGRKPVGEFGDGVDIVGWVNKTMSELSQPSD 922
YIAPEYAYTLKVDEKSDVYSFGVVLLELI GRKPVGEFGDGVDIV WV K S+
Sbjct: 2592 YIAPEYAYTLKVDEKSDVYSFGVVLLELITGRKPVGEFGDGVDIVQWVRKMTD-----SN 2756
Query: 923 TALVLAVVDPRLSGYPLTSVIHMFNIAMMCVKEMGPARPTMREVVHMLTNPPQ 975
VL V+DPRL PL V+H+F +AM+CV+E RPTMREVV +LT P+
Sbjct: 2757 KEGVLKVLDPRLPSVPLHEVMHVFYVAMLCVEEQAVERPTMREVVQILTELPK 2915
>TC219395 UP|Q9LKZ4 (Q9LKZ4) Receptor-like protein kinase 3, partial (92%)
Length = 3037
Score = 926 bits (2393), Expect = 0.0
Identities = 467/901 (51%), Positives = 623/901 (68%), Gaps = 5/901 (0%)
Frame = +3
Query: 86 LFGHLPPEIGLLEKLENLTISMNNLTDQLPSDLASLTSLKVLNISHNLFSGQFPGNITVG 145
L G L ++ L L NL+++ N + +P L++L+ L+ LN+S+N+F+ FP ++
Sbjct: 3 LSGPLSADVAHLPFLSNLSLASNKFSGPIPPSLSALSGLRFLNLSNNVFNETFPSELS-R 179
Query: 146 MTELEALDAYDNSFSGPLPEEIVKLEKLKYLHLAGNYFSGTIPESYSEFQSLEFLGLNAN 205
+ LE LD Y+N+ +G LP + +++ L++LHL GN+FSG IP Y +Q L++L ++ N
Sbjct: 180 LQNLEVLDLYNNNMTGVLPLAVAQMQNLRHLHLGGNFFSGQIPPEYGRWQRLQYLAVSGN 359
Query: 206 SLTGRVPESLAKLKTLKELHLGYSNAYEGGIPPAFGSMENLRLLEMANCNLTGEIPPSLG 265
L G +P + L +L+EL++GY N Y GGIPP G++ L L+ A C L+GEIP +LG
Sbjct: 360 ELEGTIPPEIGNLSSLRELYIGYYNTYTGGIPPEIGNLSELVRLDAAYCGLSGEIPAALG 539
Query: 266 NLTKLHSLFVQMNNLTGTIPPELSSMMSLMSLDLSINDLTGEIPESFSKLKNLTLMNFFQ 325
L KL +LF+Q+N L+G++ PEL ++ SL S+DLS N L+GEIP F +LKN+TL+N F+
Sbjct: 540 KLQKLDTLFLQVNALSGSLTPELGNLKSLKSMDLSNNMLSGEIPARFGELKNITLLNLFR 719
Query: 326 NKFRGSLPSFIGDLPNLETLQVWENNFSFVLPHNLGGNGRFLYFDVTKNHLTGLIPPDLC 385
NK G++P FIG+LP LE +Q+WENNF+ +P LG NGR D++ N LTG +P LC
Sbjct: 720 NKLHGAIPEFIGELPALEVVQLWENNFTGSIPEGLGKNGRLNLVDLSSNKLTGTLPTYLC 899
Query: 386 KSGRLKTFIITDNFFRGPIPKGIGECRSLTKIRVANNFLDGPVPPGVFQLPSVTITELSN 445
L+T I NF GPIP+ +G C SLT+IR+ NFL+G +P G+F LP +T EL +
Sbjct: 900 SGNTLQTLITLGNFLFGPIPESLGSCESLTRIRMGENFLNGSIPRGLFGLPKLTQVELQD 1079
Query: 446 NRLNGELPSVIS-GESLGTLTLSNNLFTGKIPAAMKNLRALQSLSLDANEFIGEIPGGVF 504
N L+GE P V S +LG +TLSNN +G +P ++ N ++Q L LD N F G IP +
Sbjct: 1080NYLSGEFPEVGSVAVNLGQITLSNNQLSGVLPPSIGNFSSVQKLILDGNMFTGRIPPQIG 1259
Query: 505 EIPMLTKVNISGNNLTGPIPTTITHRASLTAVDLSRNNLAGEVPKGMKNLMDLSILNLSR 564
+ L+K++ SGN +GPI I+ LT +DLSRN L+G++P + + L+ LNLSR
Sbjct: 1260RLQQLSKIDFSGNKFSGPIVPEISQCKLLTFLDLSRNELSGDIPNEITGMRILNYLNLSR 1439
Query: 565 NEISGPVPDEIRFMTSLTTLDLSSNNFTGTVPTGGQFLVFNYDKTFAGNPNLCFPHRASC 624
N + G +P I M SLT++D S NN +G VP GQF FNY +F GNP+LC P+ +C
Sbjct: 1440NHLVGGIPSSISSMQSLTSVDFSYNNLSGLVPGTGQFSYFNY-TSFLGNPDLCGPYLGAC 1616
Query: 625 PSVLYDSLRKTRAK---TARVRAIVIGIALATAVLLVAVTVHVVRKRRLHRAQAWKLTAF 681
+ + + K ++ +V+G+ L + VA ++ A+AWKLTAF
Sbjct: 1617KDGVANGAHQPHVKGLSSSFKLLLVVGLLLCSIAFAVAAIFKARSLKKASGARAWKLTAF 1796
Query: 682 QRLEIKAEDVVECLKEENIIGKGGAGIVYRGSMPNGTDVAIKRLVGQGSG-RNDYGFRAE 740
QRL+ +DV+ CLKE+NIIGKGGAGIVY+G+MPNG VA+KRL G +D+GF AE
Sbjct: 1797QRLDFTVDDVLHCLKEDNIIGKGGAGIVYKGAMPNGDHVAVKRLPAMSRGSSHDHGFNAE 1976
Query: 741 IETLGKIRHRNIMRLLGYVSNKDTNLLLYEYMPNGSLGEWLHGAKGGHLRWEMRYKIAVE 800
I+TLG+IRHR+I+RLLG+ SN +TNLL+YEYMPNGSLGE LHG KGGHL W+ RYKIAVE
Sbjct: 1977IQTLGRIRHRHIVRLLGFCSNHETNLLVYEYMPNGSLGEVLHGKKGGHLHWDTRYKIAVE 2156
Query: 801 AARGLCYMHHDCSPLIIHRDVKSNNILLDADFEAHVADFGLAKFLYDPGASQSMSSIAGS 860
AA+GLCY+HHDCSPLI+HRDVKSNNILLD++ EAHVADFGLAKFL D G S+ MS+IAGS
Sbjct: 2157AAKGLCYLHHDCSPLIVHRDVKSNNILLDSNHEAHVADFGLAKFLQDSGTSECMSAIAGS 2336
Query: 861 YGYIAPEYAYTLKVDEKSDVYSFGVVLLELIIGRKPVGEFGDGVDIVGWVNKTMSELSQP 920
YGYIAPEYAYTLKVDEKSDVYSFGVVLLELI GRKPVGEFGDGVDIV WV K
Sbjct: 2337YGYIAPEYAYTLKVDEKSDVYSFGVVLLELITGRKPVGEFGDGVDIVQWVRKMTD----- 2501
Query: 921 SDTALVLAVVDPRLSGYPLTSVIHMFNIAMMCVKEMGPARPTMREVVHMLTNPPQSNTST 980
S+ VL V+DPRL PL V+H+F +AM+CV+E RPTMREVV +LT P+ S
Sbjct: 2502SNKEGVLKVLDPRLPSVPLHEVMHVFYVAMLCVEEQAVERPTMREVVQILTELPKPPDSK 2681
Query: 981 Q 981
+
Sbjct: 2682E 2684
Score = 197 bits (501), Expect = 2e-50
Identities = 136/434 (31%), Positives = 211/434 (48%), Gaps = 7/434 (1%)
Frame = +3
Query: 75 RVVALNVTLVPLFGHLPPEIGLLEKLENLTISMNNLTDQLPSDLASLTSLKVLNISHNLF 134
R+ L V+ L G +PPEIG L L L I +N +
Sbjct: 330 RLQYLAVSGNELEGTIPPEIGNLSSLRELYIGY-----------------------YNTY 440
Query: 135 SGQFPGNITVGMTELEALDAYDNSFSGPLPEEIVKLEKLKYLHLAGNYFSGTIPESYSEF 194
+G P I ++EL LDA SG +P + KL+KL L L N SG++
Sbjct: 441 TGGIPPEIG-NLSELVRLDAAYCGLSGEIPAALGKLQKLDTLFLQVNALSGSLTPELGNL 617
Query: 195 QSLEFLGLNANSLTGRVPESLAKLKTLKELHLGYSNAYEGGIPPAFGSMENLRLLEMANC 254
+SL+ + L+ N L+G +P +LK + L+L + N G IP G + L ++++
Sbjct: 618 KSLKSMDLSNNMLSGEIPARFGELKNITLLNL-FRNKLHGAIPEFIGELPALEVVQLWEN 794
Query: 255 NLTGEIPPSLGNLTKLHSLFVQMNNLTGTIPPELSSMMSLMSLDLSINDLTGEIPESFSK 314
N TG IP LG +L+ + + N LTGT+P L S +L +L N L G IPES
Sbjct: 795 NFTGSIPEGLGKNGRLNLVDLSSNKLTGTLPTYLCSGNTLQTLITLGNFLFGPIPESLGS 974
Query: 315 LKNLTLMNFFQNKFRGSLPSFIGDLPNLETLQVWENNFSFVLPH------NLGGNGRFLY 368
++LT + +N GS+P + LP L +++ +N S P NLG
Sbjct: 975 CESLTRIRMGENFLNGSIPRGLFGLPKLTQVELQDNYLSGEFPEVGSVAVNLG------Q 1136
Query: 369 FDVTKNHLTGLIPPDLCKSGRLKTFIITDNFFRGPIPKGIGECRSLTKIRVANNFLDGPV 428
++ N L+G++PP + ++ I+ N F G IP IG + L+KI + N GP+
Sbjct: 1137ITLSNNQLSGVLPPSIGNFSSVQKLILDGNMFTGRIPPQIGRLQQLSKIDFSGNKFSGPI 1316
Query: 429 PPGVFQLPSVTITELSNNRLNGELPSVISG-ESLGTLTLSNNLFTGKIPAAMKNLRALQS 487
P + Q +T +LS N L+G++P+ I+G L L LS N G IP+++ ++++L S
Sbjct: 1317VPEISQCKLLTFLDLSRNELSGDIPNEITGMRILNYLNLSRNHLVGGIPSSISSMQSLTS 1496
Query: 488 LSLDANEFIGEIPG 501
+ N G +PG
Sbjct: 1497VDFSYNNLSGLVPG 1538
>TC210408 similar to UP|Q9FEU2 (Q9FEU2) Receptor protein kinase, partial
(15%)
Length = 994
Score = 244 bits (624), Expect = 1e-64
Identities = 127/253 (50%), Positives = 168/253 (66%), Gaps = 8/253 (3%)
Frame = +3
Query: 740 EIETLGKIRHRNIMRLLGYVSNKDTNLLLYEYMPNGSLGEWLHGAKGGHLRWEMRYKIAV 799
EI+ LG IRHRNI+R +GY SN+ NLLLY Y+PNG+L + L G + +L WE RYKIAV
Sbjct: 3 EIQILGYIRHRNIVRFIGYCSNRSINLLLYNYIPNGNLRQLLQGNR--NLDWETRYKIAV 176
Query: 800 EAARGLCYMHHDCSPLIIHRDVKSNNILLDADFEAHVADFGLAKFLYDPGASQSMSSIAG 859
+A+GL Y+HHDC P I+HRDVK NNILLD+ FEA++ADFGLAK ++ P +MS +AG
Sbjct: 177 GSAQGLAYLHHDCVPAILHRDVKCNNILLDSKFEAYLADFGLAKLMHSPNYHHAMSRVAG 356
Query: 860 SYGYIAPEYAYTLKVDEKSDVYSFGVVLLELIIGRKPV-GEFGDGVDIVGWVNKTMSELS 918
SYGYIAPEY Y++ + EKSDVYS+GVVLLE++ GR V GDG IV WV + M
Sbjct: 357 SYGYIAPEYGYSMNITEKSDVYSYGVVLLEILSGRSAVESHVGDGQHIVEWVKRKMGSFE 536
Query: 919 QPSDTALVLAVVDPRLSGYP---LTSVIHMFNIAMMCVKEMGPARPTMREVVHML----T 971
++++D +L G P + ++ IAM CV RPTM+EVV +L +
Sbjct: 537 P------AVSILDTKLQGLPDQMVQEMLQTLGIAMFCVNSSPAERPTMKEVVALLMEVKS 698
Query: 972 NPPQSNTSTQDLI 984
P + ++Q LI
Sbjct: 699 QPEEMGKTSQPLI 737
>TC217385 similar to UP|Q6ZGC7 (Q6ZGC7) Receptor protein kinase PERK1-like,
partial (80%)
Length = 1604
Score = 237 bits (605), Expect = 2e-62
Identities = 169/434 (38%), Positives = 234/434 (52%), Gaps = 27/434 (6%)
Frame = +1
Query: 580 SLTTLDLSSNNFTGTVPTGGQFLVFNYDKTFAGNPNLC-----FPHRASCPSVLYDSLRK 634
SL+ L++S N G +PT F F D +F GNP LC P + PS R
Sbjct: 4 SLSLLNVSYNKLFGVIPTSNNFTRFPPD-SFIGNPGLCGNWLNLPCHGARPSE-----RV 165
Query: 635 TRAKTARVRAIVIGIALATAVLLVAVTVHVVRKRRLHRAQAWKLTAFQR----------- 683
T +K A ++GI L V+L+ V V R H + +F +
Sbjct: 166 TLSKAA-----ILGITLGALVILLMVLVAACRP---HSPSPFPDGSFDKPINFSPPKLVI 321
Query: 684 LEIKA-----EDVV---ECLKEENIIGKGGAGIVYRGSMPNGTDVAIKRLVGQGSGRNDY 735
L + ED++ E L E+ IIG G + VY+ + N VAIKR+ +
Sbjct: 322 LHMNMALHVYEDIMRMTENLSEKYIIGYGASSTVYKCVLKNCKPVAIKRIYSHYP-QCIK 498
Query: 736 GFRAEIETLGKIRHRNIMRLLGYVSNKDTNLLLYEYMPNGSLGEWLHG-AKGGHLRWEMR 794
F E+ET+G I+HRN++ L GY + +LL Y+YM NGSL + LHG K L WE+R
Sbjct: 499 EFETELETVGSIKHRNLVSLQGYSLSPYGHLLFYDYMENGSLWDLLHGPTKKKKLDWELR 678
Query: 795 YKIAVEAARGLCYMHHDCSPLIIHRDVKSNNILLDADFEAHVADFGLAKFLYDPGASQSM 854
KIA+ AA+GL Y+HHDC P IIHRDVKS+NILLDADFE H+ DFG+AK L P S +
Sbjct: 679 LKIALGAAQGLAYLHHDCCPRIIHRDVKSSNILLDADFEPHLTDFGIAKSLC-PSKSHTS 855
Query: 855 SSIAGSYGYIAPEYAYTLKVDEKSDVYSFGVVLLELIIGRKPVGEFGDGVDIVGWVNKTM 914
+ I G+ GYI PEYA T + EKSDVYS+G+VLLEL+ GRK V + ++
Sbjct: 856 TYIMGTIGYIDPEYARTSHLTEKSDVYSYGIVLLELLTGRKAVDNESNLHHLI------- 1014
Query: 915 SELSQPSDTALVLAVVDPRLSG--YPLTSVIHMFNIAMMCVKEMGPARPTMREVVHMLTN 972
LS+ + A V+ VDP ++ L +V ++ +A++C K RPTM EV +L +
Sbjct: 1015--LSKAATNA-VMETVDPDITATCKDLGAVKKVYQLALLCTKRQPADRPTMHEVTRVLGS 1185
Query: 973 PPQSNTSTQDLINL 986
S+ + L +L
Sbjct: 1186LVPSSIPPKQLADL 1227
>TC209457 homologue to UP|Q9LKZ6 (Q9LKZ6) Receptor-like protein kinase 1,
partial (20%)
Length = 850
Score = 227 bits (579), Expect = 2e-59
Identities = 117/167 (70%), Positives = 133/167 (79%)
Frame = +2
Query: 815 LIIHRDVKSNNILLDADFEAHVADFGLAKFLYDPGASQSMSSIAGSYGYIAPEYAYTLKV 874
LI+HRDVKSNNILLD++FEAHVADFGLAKFL D GAS+ MS+IAGSYGYIAPEYAYTLKV
Sbjct: 2 LIVHRDVKSNNILLDSNFEAHVADFGLAKFLQDSGASECMSAIAGSYGYIAPEYAYTLKV 181
Query: 875 DEKSDVYSFGVVLLELIIGRKPVGEFGDGVDIVGWVNKTMSELSQPSDTALVLAVVDPRL 934
DEKSDVYSFGVVLLEL+ GRKPVGEFGDGVDIV WV K S+ VL V+DPRL
Sbjct: 182 DEKSDVYSFGVVLLELVTGRKPVGEFGDGVDIVQWVRKMTD-----SNKEGVLKVLDPRL 346
Query: 935 SGYPLTSVIHMFNIAMMCVKEMGPARPTMREVVHMLTNPPQSNTSTQ 981
PL V+H+F +AM+CV+E RPTMREVV +LT P+ +S Q
Sbjct: 347 PSVPLHEVMHVFYVAMLCVEEQAVERPTMREVVQILTELPKPPSSKQ 487
>TC209151 similar to UP|Q9FGN6 (Q9FGN6) Receptor protein kinase-like, partial
(31%)
Length = 1329
Score = 224 bits (572), Expect = 1e-58
Identities = 145/445 (32%), Positives = 231/445 (51%), Gaps = 4/445 (0%)
Frame = +1
Query: 285 PPELSSMMSLMSLDLSINDLTGEIPESFSKLKNLTLMNFFQNKFRGSLPSFIGDLPNLET 344
P ELS++ L LDLS N TG IPESFS L+NL L++ N G++P I LP+LET
Sbjct: 1 PSELSNIEPLTDLDLSDNFFTGSIPESFSDLENLRLLSVMYNDMSGTVPEGIAQLPSLET 180
Query: 345 LQVWENNFSFVLPHNLGGNGRFLYFDVTKNHLTGLIPPDLCKSGRLKTFIITDNFFRGPI 404
L +W N FS LP +LG N + + D + N L G IPPD+C SG L I+ N F G +
Sbjct: 181 LLIWNNKFSGSLPRSLGRNSKLKWVDASTNDLVGNIPPDICVSGELFKLILFSNKFTGGL 360
Query: 405 PKGIGECRSLTKIRVANNFLDGPVPPGVFQLPSVTITELSNNRLNGELPSVIS-GESLGT 463
I C SL ++R+ +N G + LP + +LS N G +PS IS L
Sbjct: 361 -SSISNCSSLVRLRLEDNLFSGEITLKFSLLPDILYVDLSRNNFVGGIPSDISQATQLEY 537
Query: 464 LTLS-NNLFTGKIPAAMKNLRALQSLSLDANEFIGEIPGGVFE-IPMLTKVNISGNNLTG 521
+S N G IP+ +L LQ+ S + ++P +FE ++ +++ N+L+G
Sbjct: 538 FNVSYNQQLGGIIPSQTWSLPQLQNFSASSCGISSDLP--LFESCKSISVIDLDSNSLSG 711
Query: 522 PIPTTITHRASLTAVDLSRNNLAGEVPKGMKNLMDLSILNLSRNEISGPVPDEIRFMTSL 581
IP ++ +L ++LS NNL G +P + ++ L +++LS N+ +GP+P + ++L
Sbjct: 712 TIPNGVSKCQALEKINLSNNNLTGHIPDELASIPVLGVVDLSNNKFNGPIPAKFGSSSNL 891
Query: 582 TTLDLSSNNFTGTVPTGGQFLVFNYDKTFAGNPNLCFPHRASCPSVLYDSLRKTRAKTAR 641
L++S NN +G++PTG F + F GN LC CP + K K R
Sbjct: 892 QLLNVSFNNISGSIPTGKSFKLMG-RSAFVGNSELCGAPLQPCPDSVGILGSKCSWKVTR 1068
Query: 642 VRAIVIGIALATAVLLVAVTVHVVRKRRLHRAQAWKLTAFQRL-EIKAEDVVECLKEENI 700
+ + +G+ ++L+ + + RR ++Q WK+ +F L + A DV+ L
Sbjct: 1069IVLLSVGL----LIVLLGLAFGMSYLRRGIKSQ-WKMVSFAGLPQFTANDVLTSLSATTK 1233
Query: 701 IGKGGAGIVYRGSMPNGTDVAIKRL 725
+ + V + +P G V +K++
Sbjct: 1234PTEVQSPSVTKAVLPTGITVLVKKI 1308
Score = 159 bits (401), Expect = 7e-39
Identities = 98/316 (31%), Positives = 168/316 (53%), Gaps = 3/316 (0%)
Frame = +1
Query: 164 PEEIVKLEKLKYLHLAGNYFSGTIPESYSEFQSLEFLGLNANSLTGRVPESLAKLKTLKE 223
P E+ +E L L L+ N+F+G+IPES+S+ ++L L + N ++G VPE +A+L +L+
Sbjct: 1 PSELSNIEPLTDLDLSDNFFTGSIPESFSDLENLRLLSVMYNDMSGTVPEGIAQLPSLET 180
Query: 224 LHLGYSNAYEGGIPPAFGSMENLRLLEMANCNLTGEIPPSLGNLTKLHSLFVQMNNLTGT 283
L L ++N + G +P + G L+ ++ + +L G IPP + +L L + N TG
Sbjct: 181 L-LIWNNKFSGSLPRSLGRNSKLKWVDASTNDLVGNIPPDICVSGELFKLILFSNKFTGG 357
Query: 284 IPPELSSMMSLMSLDLSINDLTGEIPESFSKLKNLTLMNFFQNKFRGSLPSFIGDLPNLE 343
+ +S+ SL+ L L N +GEI FS L ++ ++ +N F G +PS I LE
Sbjct: 358 L-SSISNCSSLVRLRLEDNLFSGEITLKFSLLPDILYVDLSRNNFVGGIPSDISQATQLE 534
Query: 344 TLQV-WENNFSFVLPHNLGGNGRFLYFDVTKNHLTGLIPP-DLCKSGRLKTFIITDNFFR 401
V + ++P + F + ++ +P + CKS + + N
Sbjct: 535 YFNVSYNQQLGGIIPSQTWSLPQLQNFSASSCGISSDLPLFESCKS--ISVIDLDSNSLS 708
Query: 402 GPIPKGIGECRSLTKIRVANNFLDGPVPPGVFQLPSVTITELSNNRLNGELPSVI-SGES 460
G IP G+ +C++L KI ++NN L G +P + +P + + +LSNN+ NG +P+ S +
Sbjct: 709 GTIPNGVSKCQALEKINLSNNNLTGHIPDELASIPVLGVVDLSNNKFNGPIPAKFGSSSN 888
Query: 461 LGTLTLSNNLFTGKIP 476
L L +S N +G IP
Sbjct: 889 LQLLNVSFNNISGSIP 936
Score = 143 bits (360), Expect = 4e-34
Identities = 97/317 (30%), Positives = 161/317 (50%), Gaps = 3/317 (0%)
Frame = +1
Query: 212 PESLAKLKTLKELHLGYSNAYEGGIPPAFGSMENLRLLEMANCNLTGEIPPSLGNLTKLH 271
P L+ ++ L +L L N + G IP +F +ENLRLL + +++G +P + L L
Sbjct: 1 PSELSNIEPLTDLDLS-DNFFTGSIPESFSDLENLRLLSVMYNDMSGTVPEGIAQLPSLE 177
Query: 272 SLFVQMNNLTGTIPPELSSMMSLMSLDLSINDLTGEIPESFSKLKNLTLMNFFQNKFRGS 331
+L + N +G++P L L +D S NDL G IP L + F NKF G
Sbjct: 178 TLLIWNNKFSGSLPRSLGRNSKLKWVDASTNDLVGNIPPDICVSGELFKLILFSNKFTGG 357
Query: 332 LPSFIGDLPNLETLQVWENNFSFVLPHNLGGNGRFLYFDVTKNHLTGLIPPDLCKSGRLK 391
L S I + +L L++ +N FS + LY D+++N+ G IP D+ ++ +L+
Sbjct: 358 LSS-ISNCSSLVRLRLEDNLFSGEITLKFSLLPDILYVDLSRNNFVGGIPSDISQATQLE 534
Query: 392 TFIITDN-FFRGPIPKGIGECRSLTKIRVANNFLDGPVPPGVFQ-LPSVTITELSNNRLN 449
F ++ N G IP L ++ + +P +F+ S+++ +L +N L+
Sbjct: 535 YFNVSYNQQLGGIIPSQTWSLPQLQNFSASSCGISSDLP--LFESCKSISVIDLDSNSLS 708
Query: 450 GELPSVISG-ESLGTLTLSNNLFTGKIPAAMKNLRALQSLSLDANEFIGEIPGGVFEIPM 508
G +P+ +S ++L + LSNN TG IP + ++ L + L N+F G IP
Sbjct: 709 GTIPNGVSKCQALEKINLSNNNLTGHIPDELASIPVLGVVDLSNNKFNGPIPAKFGSSSN 888
Query: 509 LTKVNISGNNLTGPIPT 525
L +N+S NN++G IPT
Sbjct: 889 LQLLNVSFNNISGSIPT 939
Score = 119 bits (297), Expect = 9e-27
Identities = 87/318 (27%), Positives = 148/318 (46%), Gaps = 1/318 (0%)
Frame = +1
Query: 115 PSDLASLTSLKVLNISHNLFSGQFPGNITVGMTELEALDAYDNSFSGPLPEEIVKLEKLK 174
PS+L+++ L L++S N F+G P + + + L L N SG +PE I +L L+
Sbjct: 1 PSELSNIEPLTDLDLSDNFFTGSIPESFS-DLENLRLLSVMYNDMSGTVPEGIAQLPSLE 177
Query: 175 YLHLAGNYFSGTIPESYSEFQSLEFLGLNANSLTGRVPESLAKLKTLKELHLGYSNAYEG 234
L + N FSG++P S L+++ + N L G +P + L +L L +SN + G
Sbjct: 178 TLLIWNNKFSGSLPRSLGRNSKLKWVDASTNDLVGNIPPDICVSGELFKLIL-FSNKFTG 354
Query: 235 GIPPAFGSMENLRLLEMANCNLTGEIPPSLGNLTKLHSLFVQMNNLTGTIPPELSSMMSL 294
G+ + + +L L + + +GEI L + + + NN G IP ++S L
Sbjct: 355 GL-SSISNCSSLVRLRLEDNLFSGEITLKFSLLPDILYVDLSRNNFVGGIPSDISQATQL 531
Query: 295 MSLDLSIN-DLTGEIPESFSKLKNLTLMNFFQNKFRGSLPSFIGDLPNLETLQVWENNFS 353
++S N L G IP L L + LP F ++ + + N+ S
Sbjct: 532 EYFNVSYNQQLGGIIPSQTWSLPQLQNFSASSCGISSDLPLF-ESCKSISVIDLDSNSLS 708
Query: 354 FVLPHNLGGNGRFLYFDVTKNHLTGLIPPDLCKSGRLKTFIITDNFFRGPIPKGIGECRS 413
+P+ + +++ N+LTG IP +L L +++N F GPIP G +
Sbjct: 709 GTIPNGVSKCQALEKINLSNNNLTGHIPDELASIPVLGVVDLSNNKFNGPIPAKFGSSSN 888
Query: 414 LTKIRVANNFLDGPVPPG 431
L + V+ N + G +P G
Sbjct: 889 LQLLNVSFNNISGSIPTG 942
Score = 96.7 bits (239), Expect = 5e-20
Identities = 62/195 (31%), Positives = 97/195 (48%)
Frame = +1
Query: 88 GHLPPEIGLLEKLENLTISMNNLTDQLPSDLASLTSLKVLNISHNLFSGQFPGNITVGMT 147
G + + LL + + +S NN +PSD++ T L+ N+S+N G + T +
Sbjct: 421 GEITLKFSLLPDILYVDLSRNNFVGGIPSDISQATQLEYFNVSYNQQLGGIIPSQTWSLP 600
Query: 148 ELEALDAYDNSFSGPLPEEIVKLEKLKYLHLAGNYFSGTIPESYSEFQSLEFLGLNANSL 207
+L+ A S LP + + + L N SGTIP S+ Q+LE + L+ N+L
Sbjct: 601 QLQNFSASSCGISSDLP-LFESCKSISVIDLDSNSLSGTIPNGVSKCQALEKINLSNNNL 777
Query: 208 TGRVPESLAKLKTLKELHLGYSNAYEGGIPPAFGSMENLRLLEMANCNLTGEIPPSLGNL 267
TG +P+ LA + L + L +N + G IP FGS NL+LL ++ N++G IP
Sbjct: 778 TGHIPDELASIPVLGVVDLS-NNKFNGPIPAKFGSSSNLQLLNVSFNNISGSIPTGKSFK 954
Query: 268 TKLHSLFVQMNNLTG 282
S FV + L G
Sbjct: 955 LMGRSAFVGNSELCG 999
Score = 53.1 bits (126), Expect = 6e-07
Identities = 32/119 (26%), Positives = 59/119 (48%), Gaps = 5/119 (4%)
Frame = +1
Query: 51 EDWKFSTSLSAHCSFSGVTCDQNLRVVALNVTLVPL-----FGHLPPEIGLLEKLENLTI 105
+ W + S G++ D L +++++ L G +P + + LE + +
Sbjct: 583 QTWSLPQLQNFSASSCGISSDLPLFESCKSISVIDLDSNSLSGTIPNGVSKCQALEKINL 762
Query: 106 SMNNLTDQLPSDLASLTSLKVLNISHNLFSGQFPGNITVGMTELEALDAYDNSFSGPLP 164
S NNLT +P +LAS+ L V+++S+N F+G P + L+ L+ N+ SG +P
Sbjct: 763 SNNNLTGHIPDELASIPVLGVVDLSNNKFNGPIPAKFG-SSSNLQLLNVSFNNISGSIP 936
>TC216858 homologue to UP|Q76FZ8 (Q76FZ8) Brassinosteroid receptor, partial
(29%)
Length = 1320
Score = 218 bits (555), Expect = 1e-56
Identities = 123/288 (42%), Positives = 179/288 (61%), Gaps = 8/288 (2%)
Frame = +1
Query: 690 DVVECLKEENIIGKGGAGIVYRGSMPNGTDVAIKRLVGQGSGRNDYGFRAEIETLGKIRH 749
D +++IG GG G VY+ + +G+ VAIK+L+ SG+ D F AE+ET+GKI+H
Sbjct: 67 DATNGFHNDSLIGSGGFGDVYKAQLKDGSVVAIKKLI-HVSGQGDREFTAEMETIGKIKH 243
Query: 750 RNIMRLLGYVSNKDTNLLLYEYMPNGSLGEWLHGAK--GGHLRWEMRYKIAVEAARGLCY 807
RN++ LLGY + LL+YEYM GSL + LH K G L W +R KIA+ AARGL +
Sbjct: 244 RNLVPLLGYCKVGEERLLVYEYMKYGSLEDVLHDQKKAGIKLNWAIRRKIAIGAARGLAF 423
Query: 808 MHHDCSPLIIHRDVKSNNILLDADFEAHVADFGLAKFLYDPGASQSMSSIAGSYGYIAPE 867
+HH+C P IIHRD+KS+N+LLD + EA V+DFG+A+ + S+S++AG+ GY+ PE
Sbjct: 424 LHHNCIPHIIHRDMKSSNVLLDENLEARVSDFGMARLMSAMDTHLSVSTLAGTPGYVPPE 603
Query: 868 YAYTLKVDEKSDVYSFGVVLLELIIGRKPV--GEFGDGVDIVGWVNK----TMSELSQPS 921
Y + + K DVYS+GVVLLEL+ G++P +FGD ++VGWV + +S++ P
Sbjct: 604 YYQSFRCSTKGDVYSYGVVLLELLTGKRPTDSADFGDN-NLVGWVKQHAKLKISDIFDPE 780
Query: 922 DTALVLAVVDPRLSGYPLTSVIHMFNIAMMCVKEMGPARPTMREVVHM 969
L DP L ++ IA+ C+ + RPTM +V+ M
Sbjct: 781 -----LMKEDPNLE----MELLQHLKIAVSCLDDRPWRRPTMIQVMAM 897
>TC218252 weakly similar to UP|Q03407 (Q03407) Ydr490cp, partial (5%)
Length = 1279
Score = 206 bits (523), Expect = 5e-53
Identities = 111/252 (44%), Positives = 161/252 (63%), Gaps = 2/252 (0%)
Frame = +1
Query: 721 AIKRLVGQGSGRNDYGFRAEIETLGKIRHRNIMRLLGYVSNKDTNLLLYEYMPNGSLGEW 780
A+KR+V G + + F E+E LG I+HR ++ L GY ++ + LL+Y+Y+P GSL E
Sbjct: 1 ALKRIVKLNEGFDRF-FERELEILGSIKHRYLVNLRGYCNSPTSKLLIYDYLPGGSLDEA 177
Query: 781 LHGAKGGHLRWEMRYKIAVEAARGLCYMHHDCSPLIIHRDVKSNNILLDADFEAHVADFG 840
LH + L W+ R I + AA+GL Y+HHDCSP IIHRD+KS+NILLD + EA V+DFG
Sbjct: 178 LH-ERADQLDWDSRLNIIMGAAKGLAYLHHDCSPRIIHRDIKSSNILLDGNLEARVSDFG 354
Query: 841 LAKFLYDPGASQSMSSIAGSYGYIAPEYAYTLKVDEKSDVYSFGVVLLELIIGRKPVGE- 899
LAK L D S + +AG++GY+APEY + + EKSDVYSFGV+ LE++ G++P
Sbjct: 355 LAKLLEDE-ESHITTIVAGTFGYLAPEYMQSGRATEKSDVYSFGVLTLEVLSGKRPTDAA 531
Query: 900 -FGDGVDIVGWVNKTMSELSQPSDTALVLAVVDPRLSGYPLTSVIHMFNIAMMCVKEMGP 958
G +IVGW+N ++E ++P + +VDP G + S+ + ++A+ CV
Sbjct: 532 FIEKGPNIVGWLNFLITE-NRPRE------IVDPLCEGVQMESLDALLSVAIQCVSSSPE 690
Query: 959 ARPTMREVVHML 970
RPTM VV +L
Sbjct: 691 DRPTMHRVVQLL 726
>TC208867 similar to UP|Q9LXT2 (Q9LXT2) Serine/threonine-specific protein
kinase-like protein, partial (72%)
Length = 1118
Score = 205 bits (521), Expect = 9e-53
Identities = 122/283 (43%), Positives = 166/283 (58%), Gaps = 9/283 (3%)
Frame = +1
Query: 697 EENIIGKGGAGIVYRGSMPNGTDVAIKRLVGQGSGRNDYGFRAEIETLGKIRHRNIMRLL 756
+ N+IG GG G+VYRG + +G VAIK + Q + + F+ E+E L ++ ++ LL
Sbjct: 4 KSNVIGHGGFGLVYRGVLNDGRKVAIK-FMDQAGKQGEEEFKVEVELLSRLHSPYLLALL 180
Query: 757 GYVSNKDTNLLLYEYMPNGSLGEWLHGAKGG-----HLRWEMRYKIAVEAARGLCYMHHD 811
GY S+ + LL+YE+M NG L E L+ L WE R +IA+EAA+GL Y+H
Sbjct: 181 GYCSDSNHKLLVYEFMANGGLQEHLYPVSNSIITPVKLDWETRLRIALEAAKGLEYLHEH 360
Query: 812 CSPLIIHRDVKSNNILLDADFEAHVADFGLAKFLYDPGASQSMSSIAGSYGYIAPEYAYT 871
SP +IHRD KS+NILLD F A V+DFGLAK D + + G+ GY+APEYA T
Sbjct: 361 VSPPVIHRDFKSSNILLDKKFHAKVSDFGLAKLGPDRAGGHVSTRVLGTQGYVAPEYALT 540
Query: 872 LKVDEKSDVYSFGVVLLELIIGRKPVG---EFGDGVDIVGWVNKTMSELSQPSDTALVLA 928
+ KSDVYS+GVVLLEL+ GR PV G+GV +V W L +D V+
Sbjct: 541 GHLTTKSDVYSYGVVLLELLTGRVPVDMKRPPGEGV-LVSWA------LPLLTDREKVVK 699
Query: 929 VVDPRLSG-YPLTSVIHMFNIAMMCVKEMGPARPTMREVVHML 970
++DP L G Y + V+ + IA MCV+ RP M +VV L
Sbjct: 700 IMDPSLEGQYSMKEVVQVAAIAAMCVQPEADYRPLMADVVQSL 828
>CO979898
Length = 869
Score = 204 bits (520), Expect = 1e-52
Identities = 109/221 (49%), Positives = 147/221 (66%), Gaps = 3/221 (1%)
Frame = -1
Query: 695 LKEENIIGKGGAGIVYRGSMPNGTDVAIKRLVGQGSGRNDYGFRAEIETLGKIRHRNIMR 754
L ++IIG GG G+VY + + T +AIKRL +G+ D GF E+E + I+HRNI+
Sbjct: 863 LNSKDIIGSGGYGVVYELKLDDSTALAIKRL-NRGTAERDKGFERELEAMADIKHRNIVT 687
Query: 755 LLGYVSNKDTNLLLYEYMPNGSLGEWLHG-AKGGHLRWEMRYKIAVEAARGLCYMHHDCS 813
L GY + NLL+YE MP+GSL +LHG ++ L W RY+IA AARG+ Y+HHDC
Sbjct: 686 LHGYYTAPLYNLLIYELMPHGSLDSFLHGRSREKVLDWPTRYRIAAGAARGISYLHHDCI 507
Query: 814 PLIIHRDVKSNNILLDADFEAHVADFGLAKFLYDPGASQSMSSIAGSYGYIAPEYAYTLK 873
P IIHRD+KS+NILLD + +A V+DFGLA L P + + +AG++GY+APEY T +
Sbjct: 506 PHIIHRDIKSSNILLDQNMDARVSDFGLAT-LMQPNKTHVSTIVAGTFGYLAPEYFDTGR 330
Query: 874 VDEKSDVYSFGVVLLELIIGRKPVGE--FGDGVDIVGWVNK 912
K DVYSFGVVLLEL+ G+KP E +G +V WV +
Sbjct: 329 ATLKGDVYSFGVVLLELLTGKKPSDEAFMEEGTMLVTWVRQ 207
>TC206120 similar to UP|Q941F6 (Q941F6) Leucine-rich repeat receptor-like
kinase F21M12.36, partial (18%)
Length = 1037
Score = 199 bits (506), Expect = 5e-51
Identities = 109/230 (47%), Positives = 148/230 (63%), Gaps = 3/230 (1%)
Frame = +3
Query: 744 LGKIRHRNIMRLLGYVSNKDTNLLLYEYMPNGSLGEWLHGAKGGHLRWEMRYKIAVEAAR 803
L IRH N+++L ++++D++LL+YEY+PNGSL + LH ++ L WE RY+IAV AA+
Sbjct: 3 LSSIRHVNVVKLFCSITSEDSSLLVYEYLPNGSLWDRLHTSRKMELDWETRYEIAVGAAK 182
Query: 804 GLCYMHHDCSPLIIHRDVKSNNILLDADFEAHVADFGLAKFLYDPGASQSMSS-IAGSYG 862
GL Y+HH C +IHRDVKS+NILLD + +ADFGLAK + S + IAG++G
Sbjct: 183 GLEYLHHGCEKPVIHRDVKSSNILLDEFLKPRIADFGLAKVIQANVVKDSSTHVIAGTHG 362
Query: 863 YIAPEYAYTLKVDEKSDVYSFGVVLLELIIGRKPV-GEFGDGVDIVGWV-NKTMSELSQP 920
YIAPEY YT KV+EKSDVYSFGVVL+EL+ G++P EFG+ DIV WV NK S+
Sbjct: 363 YIAPEYGYTYKVNEKSDVYSFGVVLMELVTGKRPTEPEFGENKDIVSWVHNKARSKEG-- 536
Query: 921 SDTALVLAVVDPRLSGYPLTSVIHMFNIAMMCVKEMGPARPTMREVVHML 970
+ + VD R+ + A++C + RPTMR VV L
Sbjct: 537 -----LRSAVDSRIPEMYTEEACKVLRTAVLCTGTLPALRPTMRAVVQKL 671
>TC229875 weakly similar to UP|Q75WU3 (Q75WU3) Leucine-rich repeat
receptor-like protein kinase 1, partial (23%)
Length = 1141
Score = 198 bits (504), Expect = 9e-51
Identities = 118/301 (39%), Positives = 181/301 (59%), Gaps = 8/301 (2%)
Frame = +2
Query: 690 DVVECLKEENIIGKGGAGIVYRGSMPNGTDVAIKRL--VGQGSGRNDYGFRAEIETLGKI 747
+ E ++++IG GG G VY+ +P G VA+K+L V G N F EI+ L +I
Sbjct: 35 EATEDFDDKHLIGVGGQGCVYKAVLPTGQVVAVKKLHSVPNGEMLNLKAFTCEIQALTEI 214
Query: 748 RHRNIMRLLGYVSNKDTNLLLYEYMPNGSLGEWLHG-AKGGHLRWEMRYKIAVEAARGLC 806
RHRNI++L G+ S+ + L+ E++ NGS+G+ L + W R + + A LC
Sbjct: 215 RHRNIVKLYGFCSHSQFSFLVCEFLENGSVGKTLKDDGQAMAFDWYKRVNVVKDVANALC 394
Query: 807 YMHHDCSPLIIHRDVKSNNILLDADFEAHVADFGLAKFLYDPGASQSMSSIAGSYGYIAP 866
YMHH+CSP I+HRD+ S N+LLD+++ AHV+DFG AKFL +P +S + +S G++GY AP
Sbjct: 395 YMHHECSPRIVHRDISSKNVLLDSEYVAHVSDFGTAKFL-NPDSS-NWTSFVGTFGYAAP 568
Query: 867 EYAYTLKVDEKSDVYSFGVVLLELIIGRKPVGEFGDGVD-IVGWVNKTMSELSQPSDTAL 925
E AYT++V+EK DVYSFGV+ E++IG+ P GD + ++G T+ ++ D
Sbjct: 569 ELAYTMEVNEKCDVYSFGVLAWEILIGKHP----GDVISCLLGSSPSTL--VASTLDHMA 730
Query: 926 VLAVVDPRLSGYPL----TSVIHMFNIAMMCVKEMGPARPTMREVVHMLTNPPQSNTSTQ 981
++ +D RL +P V + IAM C+ E +RPTM +V + L S++S+
Sbjct: 731 LMDKLDQRLP-HPTKPIGKEVASIAKIAMACLTESPRSRPTMEQVANELV---MSSSSSM 898
Query: 982 D 982
D
Sbjct: 899 D 901
>TC212439 similar to UP|Q9LVP0 (Q9LVP0) Receptor-like protein kinase, partial
(15%)
Length = 491
Score = 194 bits (492), Expect = 2e-49
Identities = 97/165 (58%), Positives = 120/165 (71%), Gaps = 1/165 (0%)
Frame = +2
Query: 703 KGGAGIVYRGSMPNGTDVAIKRLVGQGSGRN-DYGFRAEIETLGKIRHRNIMRLLGYVSN 761
KG G VY+ M +G +A+K+L G N + FRAEI TLG+IRHRNI++L G+
Sbjct: 2 KGACGTVYKAMMKSGKTIAVKKLASNREGNNIENSFRAEITTLGRIRHRNIVKLYGFCYQ 181
Query: 762 KDTNLLLYEYMPNGSLGEWLHGAKGGHLRWEMRYKIAVEAARGLCYMHHDCSPLIIHRDV 821
+ +NLLLYEYM GSLGE LHG +L W +R+ IA+ AA GL Y+HHDC P IIHRD+
Sbjct: 182 QGSNLLLYEYMERGSLGELLHG-NASNLEWPIRFMIALGAAEGLAYLHHDCKPKIIHRDI 358
Query: 822 KSNNILLDADFEAHVADFGLAKFLYDPGASQSMSSIAGSYGYIAP 866
KSNNILLD +FEAHV DFGLAK + D S+SMS++AGSYGYIAP
Sbjct: 359 KSNNILLDENFEAHVGDFGLAKVI-DMPQSKSMSAVAGSYGYIAP 490
>BU760718 similar to GP|3641252|gb| leucine-rich receptor-like protein kinase
{Malus x domestica}, partial (14%)
Length = 445
Score = 190 bits (482), Expect = 3e-48
Identities = 95/148 (64%), Positives = 113/148 (76%), Gaps = 1/148 (0%)
Frame = +1
Query: 728 QGSGRNDYGFRAEIETLGKIRHRNIMRLLGYVSNKDTNLLLYEYMPNGSLGEWLHGAKGG 787
+G D F AE+ETLGKIRH+NI++L + +D LL+YEYMPNGSLG+ LH +KGG
Sbjct: 1 KGGRVQDNAFDAEVETLGKIRHKNIVKLWCCCTTRDCKLLVYEYMPNGSLGDLLHSSKGG 180
Query: 788 HLRWEMRYKIAVEAARGLCYMHHDCSPLIIHRDVKSNNILLDADFEAHVADFGLAKFL-Y 846
L W RYKIAV+AA GL Y+HHDC P I+HRDVKSNNILLD DF A VADFG+AK +
Sbjct: 181 LLDWPTRYKIAVDAAEGLSYLHHDCVPAIVHRDVKSNNILLDVDFGARVADFGVAKAVET 360
Query: 847 DPGASQSMSSIAGSYGYIAPEYAYTLKV 874
P ++SMS IAGS GYIAPEYAYTL+V
Sbjct: 361 TPKGAKSMSVIAGSCGYIAPEYAYTLRV 444
>TC209224 similar to UP|BRI1_LYCPE (Q8L899) Systemin receptor SR160 precursor
(Brassinosteroid LRR receptor kinase) , partial (14%)
Length = 880
Score = 189 bits (481), Expect = 4e-48
Identities = 107/241 (44%), Positives = 148/241 (61%), Gaps = 9/241 (3%)
Frame = +3
Query: 739 AEIETLGKIRHRNIMRLLGYVSNKDTNLLLYEYMPNGSLGEWLHGAKGGH----LRWEMR 794
AE+ETLGKI+HRN++ LLGY + LL+YEYM GSL E LHG L WE R
Sbjct: 3 AEMETLGKIKHRNLVPLLGYCKVGEERLLVYEYMEYGSLEEMLHGRIKTRDRRILTWEER 182
Query: 795 YKIAVEAARGLCYMHHDCSPLIIHRDVKSNNILLDADFEAHVADFGLAKFLYDPGASQSM 854
KIA AA+GLC++HH+C P IIHRD+KS+N+LLD + E+ V+DFG+A+ + S+
Sbjct: 183 KKIARGAAKGLCFLHHNCIPHIIHRDMKSSNVLLDHEMESRVSDFGMARLISALDTHLSV 362
Query: 855 SSIAGSYGYIAPEYAYTLKVDEKSDVYSFGVVLLELIIGRKPVG--EFGDGVDIVGWVNK 912
S++AG+ GY+ PEY + + K DVYSFGVV+LEL+ G++P +FGD ++VGW
Sbjct: 363 STLAGTPGYVPPEYYQSFRCTVKGDVYSFGVVMLELLSGKRPTDKEDFGD-TNLVGWAKI 539
Query: 913 TMSELSQPS--DTALVLAVV-DPRLSGYPLTSVIHMFNIAMMCVKEMGPARPTMREVVHM 969
+ E Q D L+LA + +I I + CV ++ RP M +VV M
Sbjct: 540 KVREGKQMEVIDNDLLLATQGTDEAEAKEVKEMIRYLEITLQCVDDLPSRRPNMLQVVAM 719
Query: 970 L 970
L
Sbjct: 720 L 722
>TC204549 similar to UP|Q9M0X5 (Q9M0X5) Receptor protein kinase-like protein,
partial (54%)
Length = 2032
Score = 188 bits (477), Expect = 1e-47
Identities = 125/349 (35%), Positives = 185/349 (52%), Gaps = 12/349 (3%)
Frame = +3
Query: 644 AIVIGIALATAVLLVAVTVHVVRKRRLHRAQ------AWKLTAFQRLEIK---AEDVVEC 694
AIV+ I +A + +V + R R+ + A+ + L+ E
Sbjct: 648 AIVVPITVAVLIFIVGICFLSRRARKKQQGSVKEGKTAYDIPTVDSLQFDFSTIEAATNK 827
Query: 695 LKEENIIGKGGAGIVYRGSMPNGTDVAIKRLVGQGSGRNDYGFRAEIETLGKIRHRNIMR 754
+N +G+GG G VY+G++ +G VA+KRL + SG+ F+ E+ + K++HRN++R
Sbjct: 828 FSADNKLGEGGFGEVYKGTLSSGQVVAVKRL-SKSSGQGGEEFKNEVVVVAKLQHRNLVR 1004
Query: 755 LLGYVSNKDTNLLLYEYMPNGSLGEWLHGA-KGGHLRWEMRYKIAVEAARGLCYMHHDCS 813
LLG+ + +L+YEY+PN SL L K L W RYKI ARG+ Y+H D
Sbjct: 1005LLGFCLQGEEKILVYEYVPNKSLDYILFDPEKQRELDWGRRYKIIGGIARGIQYLHEDSR 1184
Query: 814 PLIIHRDVKSNNILLDADFEAHVADFGLAKFLYDPGASQSMSSIAGSYGYIAPEYAYTLK 873
IIHRD+K++NILLD D ++DFG+A+ + S I G+YGY+APEYA +
Sbjct: 1185LRIIHRDLKASNILLDGDMNPKISDFGMARIFGVDQTQGNTSRIVGTYGYMAPEYAMHGE 1364
Query: 874 VDEKSDVYSFGVVLLELIIGRKPVGEF-GDGVDIVGWVNKTMSELSQPSDTALVLAVVDP 932
KSDVYSFGV+L+E++ G+K + DG + +S Q L ++DP
Sbjct: 1365FSVKSDVYSFGVLLMEILSGKKNSSFYQTDGAE------DLLSYAWQLWKDGTPLELMDP 1526
Query: 933 RL-SGYPLTSVIHMFNIAMMCVKEMGPARPTMREVVHMLTNPPQSNTST 980
L Y VI +I ++CV+E RPTM +V ML SNT T
Sbjct: 1527ILRESYNQNEVIRSIHIGLLCVQEDPADRPTMATIVLML----DSNTVT 1661
>TC230014 similar to GB|AAP21294.1|30102752|BT006486 At5g49760 {Arabidopsis
thaliana;} , partial (29%)
Length = 1087
Score = 187 bits (475), Expect = 2e-47
Identities = 106/260 (40%), Positives = 155/260 (58%), Gaps = 1/260 (0%)
Frame = +3
Query: 709 VYRGSMPNGTDVAIKRLVGQGSGRNDYGFRAEIETLGKIRHRNIMRLLGYVSNKDTNLLL 768
VY+G++P+G VAIKR + S + F+ EIE L ++ H+N++ L+G+ K +L+
Sbjct: 6 VYQGTLPSGELVAIKRAAKE-SMQGAVEFKTEIELLSRVHHKNLVGLVGFCFEKGEQMLV 182
Query: 769 YEYMPNGSLGEWLHGAKGGHLRWEMRYKIAVEAARGLCYMHHDCSPLIIHRDVKSNNILL 828
YE++PNG+L + L G G + W R K+A+ AARGL Y+H P IIHRD+KS+NILL
Sbjct: 183 YEHIPNGTLMDSLSGKSGIWMDWIRRLKVALGAARGLAYLHELADPPIIHRDIKSSNILL 362
Query: 829 DADFEAHVADFGLAKFLYDPGASQSMSSIAGSYGYIAPEYAYTLKVDEKSDVYSFGVVLL 888
D A VADFGL+K L D + + G+ GY+ PEY T ++ EKSDVYS+GV++L
Sbjct: 363 DHHLNAKVADFGLSKLLVDSERGHVTTQVKGTMGYLDPEYYMTQQLTEKSDVYSYGVLML 542
Query: 889 ELIIGRKPVGEFGDGVDIVGWVNKTMSELSQPSDTALVLAVVDPRLSGYPLTSVIHMF-N 947
EL R+P+ E G + V + M + D + ++++P + + F
Sbjct: 543 ELATARRPI-EQGKYI-----VREVMRVMDTSKDLYNLHSILNPTIMKATRPKGLEKFVM 704
Query: 948 IAMMCVKEMGPARPTMREVV 967
+AM CVKE RPTM EVV
Sbjct: 705 LAMRCVKEYAAERPTMAEVV 764
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.319 0.138 0.403
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 41,725,348
Number of Sequences: 63676
Number of extensions: 590615
Number of successful extensions: 7060
Number of sequences better than 10.0: 1154
Number of HSP's better than 10.0 without gapping: 4056
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 5351
length of query: 986
length of database: 12,639,632
effective HSP length: 106
effective length of query: 880
effective length of database: 5,889,976
effective search space: 5183178880
effective search space used: 5183178880
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 64 (29.3 bits)
Lotus: description of TM0258c.2


