

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0230.6
(1145 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
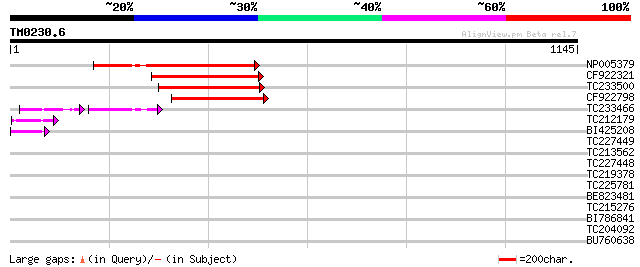
Score E
Sequences producing significant alignments: (bits) Value
NP005379 ORF (334 AA) 348 8e-96
CF922321 226 6e-59
TC233500 similar to UP|Q6I8M7 (Q6I8M7) Soybean Tgm5 transposable... 206 6e-53
CF922798 179 8e-45
TC233466 similar to UP|Q6I8M7 (Q6I8M7) Soybean Tgm5 transposable... 119 7e-37
TC212179 63 8e-10
BI425208 55 1e-07
TC227449 similar to UP|Q8S902 (Q8S902) Syringolide-induced prote... 36 0.11
TC213562 similar to UP|Q70US4 (Q70US4) NADH dehydrogenase 6, par... 34 0.42
TC227448 homologue to UP|Q8S902 (Q8S902) Syringolide-induced pro... 32 1.6
TC219378 similar to UP|SLY1_ARATH (Q9SL48) SEC1-family transport... 31 2.7
TC225781 similar to GB|AAK11228.1|12963361|AF327524 copper-bindi... 30 4.6
BE823481 30 4.6
TC215276 UP|143C_SOYBN (Q96452) 14-3-3-like protein C (SGF14C), ... 30 6.1
BI786841 30 7.9
TC204092 homologue to UP|PSAD_NICSY (P29302) Photosystem I react... 30 7.9
BU760638 weakly similar to GP|23237921|db contains ESTs D41667(S... 30 7.9
>NP005379 ORF (334 AA)
Length = 1002
Score = 348 bits (893), Expect = 8e-96
Identities = 162/335 (48%), Positives = 223/335 (66%)
Frame = +1
Query: 169 HGWSDKSFSMLLDLLRDAFPEGNVLPKSFYETKKMISELGLGYEKIHACPNDCILYWDKY 228
+GWSDKSF+ LL ++ D P+ N LPKS+Y+ KK++ +G+ Y+KIHACPNDCILY ++
Sbjct: 1 YGWSDKSFTSLLQIVHDLLPQDNTLPKSYYQAKKILCPMGMEYQKIHACPNDCILYRHEF 180
Query: 229 TNDEVCPKCGASRWKTVTEEVQPNGTKTSEKQKNTPLPKSVPLPGIPAKILRWFPLKPRL 288
CP+CGASR+K +E + E K P PAK+L + P+ PR
Sbjct: 181 EEMSKCPRCGASRYKVKDDE----DCSSDENSKKGP----------PAKVLWYLPIIPRF 318
Query: 289 QRLFMSSKIAEAMRWHRDGRLDDGFLRHPADSLAWKKFDSKYQTFALDPHNVRLGLASDG 348
+RLF + A+ + WH +GR DG +RHPAD WKK DS Y F + N+RLGLASDG
Sbjct: 319 KRLFANEDDAKDLTWHANGRKSDGMVRHPADCSQWKKIDSLYPNFGKEARNLRLGLASDG 498
Query: 349 FNPFKTMSITHST*PVILVHFNLPPWMCMKQSFFMLSLLIPGPKGPGNNIDTYLQPLVKE 408
NP+ +S HS+ PV+LV +N PPW+CMK+ + MLS++I GP+ PGN+ID YL PL+++
Sbjct: 499 MNPYGNLSTQHSSWPVLLVIYNFPPWLCMKRKYMMLSMMISGPRQPGNDIDVYLSPLIED 678
Query: 409 LQELWENGLETFDAYKKESFQLHAALMWTINDFPAYANLSGWSTKGKYACPCCGFETSSQ 468
L++LW+ G+ FD ++KE+FQ+ A L TINDFPAY NLSG+S KG ACP C +TS
Sbjct: 679 LRKLWDEGVLVFDGFRKETFQMRAMLFCTINDFPAYGNLSGYSVKGHLACPICEEDTSYI 858
Query: 469 WLRNGRKFCYMSHRRWLTPNHKWRLNGKDFDGTQE 503
L++GRK Y HR +L +H +R K F+G+QE
Sbjct: 859 QLKHGRKTVYTRHRVFLKAHHPYRRLKKAFNGSQE 963
>CF922321
Length = 742
Score = 226 bits (575), Expect = 6e-59
Identities = 109/226 (48%), Positives = 148/226 (65%)
Frame = -1
Query: 286 PRLQRLFMSSKIAEAMRWHRDGRLDDGFLRHPADSLAWKKFDSKYQTFALDPHNVRLGLA 345
PRL+RLF + A+ + WH +GR DG +R A S WKK D Y + N+RLGLA
Sbjct: 736 PRLKRLFANGDDAKDLTWHANGRNCDGMIRRLAVSSQWKKIDRLYLDLGKEASNLRLGLA 557
Query: 346 SDGFNPFKTMSITHST*PVILVHFNLPPWMCMKQSFFMLSLLIPGPKGPGNNIDTYLQPL 405
SDG NP+ + S HS+ PV+LV +NLPPW+CMK+ + MLS++I P+ PGN+ID YL L
Sbjct: 556 SDGMNPYGSSST*HSSWPVLLVIYNLPPWLCMKRKYMMLSMMISNPRQPGNDIDVYLSSL 377
Query: 406 VKELQELWENGLETFDAYKKESFQLHAALMWTINDFPAYANLSGWSTKGKYACPCCGFET 465
+++L +LW+ G F+ + E+F + A L TINDFPAY NLSG++ KG ++CP C T
Sbjct: 376 IEDLTKLWDEGFLAFNGFWNETFPMRAMLFCTINDFPAYGNLSGYNVKGHHSCPICEENT 197
Query: 466 SSQWLRNGRKFCYMSHRRWLTPNHKWRLNGKDFDGTQEFRDPPIRL 511
S L++GRK Y HRR+LTPNH +R K F+G+Q PI L
Sbjct: 196 SYIQLKHGRKTVYSRHRRFLTPNHPYRRLKKAFNGSQGHDIVPIPL 59
>TC233500 similar to UP|Q6I8M7 (Q6I8M7) Soybean Tgm5 transposable element
(Fragment), partial (65%)
Length = 1033
Score = 206 bits (523), Expect = 6e-53
Identities = 99/213 (46%), Positives = 138/213 (64%)
Frame = +2
Query: 301 MRWHRDGRLDDGFLRHPADSLAWKKFDSKYQTFALDPHNVRLGLASDGFNPFKTMSITHS 360
+ WH +GR D LRH ADSL WKK D Y F + N+ LGLA+D NP+ +S H+
Sbjct: 8 LTWHANGRNCDRMLRHLADSLQWKKIDRLYPDFGKETKNLTLGLATDAMNPYGCLSTQHN 187
Query: 361 T*PVILVHFNLPPWMCMKQSFFMLSLLIPGPKGPGNNIDTYLQPLVKELQELWENGLETF 420
+ PV+LV +NLP W+CMK+ + MLS++I GP+ PGN+I+ YL PL+++L +L + G+ F
Sbjct: 188 SWPVLLVIYNLPHWLCMKRKYMMLSMMITGPRQPGNDINVYLNPLIEDLTKL*DEGVLVF 367
Query: 421 DAYKKESFQLHAALMWTINDFPAYANLSGWSTKGKYACPCCGFETSSQWLRNGRKFCYMS 480
D ++ E+F L A L TINDFP Y NLSG+S KG ACP C +TS ++GRK Y
Sbjct: 368 DGFRNETFNLCAMLFCTINDFPTYENLSGYSVKGHRACPICEEDTSYIQQKHGRKTVYTR 547
Query: 481 HRRWLTPNHKWRLNGKDFDGTQEFRDPPIRLDG 513
HRR+L P+H +R K F+ + E + + L G
Sbjct: 548 HRRFLKPHHPYRRLKKAFNES*EHENALVLLTG 646
>CF922798
Length = 617
Score = 179 bits (453), Expect = 8e-45
Identities = 88/197 (44%), Positives = 127/197 (63%)
Frame = -2
Query: 327 DSKYQTFALDPHNVRLGLASDGFNPFKTMSITHST*PVILVHFNLPPWMCMKQSFFMLSL 386
D Y F+ + N+RLGLA+DG NP+ ++S +S V+LV +NL PW+CM + + MLS+
Sbjct: 613 DHLYLDFSKEAINLRLGLATDGMNPYGSLSTQYSLWSVLLVIYNLSPWLCMNKKYMMLSM 434
Query: 387 LIPGPKGPGNNIDTYLQPLVKELQELWENGLETFDAYKKESFQLHAALMWTINDFPAYAN 446
+I P+ PGN+ID YL PL+++L L + + FD ++ E+F L A L TINDFPAY N
Sbjct: 433 MILSPRQPGNDIDVYLSPLIEDLTML*DEEVLVFDGFQNETFHLCAMLFCTINDFPAYGN 254
Query: 447 LSGWSTKGKYACPCCGFETSSQWLRNGRKFCYMSHRRWLTPNHKWRLNGKDFDGTQEFRD 506
LSG+S K ACP C +TS L++GRK Y H+R+L P+H +R K+F+G+QE
Sbjct: 253 LSGYSVKSHRACPICEEDTSYIQLKHGRKTIYTRHQRFLKPHHLYRRLKKEFNGSQEHET 74
Query: 507 PPIRLDGTAQLRQLDEI 523
PI L G +Q+ +
Sbjct: 73 TPILLVGDHVYQQVQHL 23
>TC233466 similar to UP|Q6I8M7 (Q6I8M7) Soybean Tgm5 transposable element
(Fragment), partial (37%)
Length = 790
Score = 119 bits (297), Expect(2) = 7e-37
Identities = 59/149 (39%), Positives = 87/149 (57%)
Frame = +2
Query: 159 MVHLYHLKCLHGWSDKSFSMLLDLLRDAFPEGNVLPKSFYETKKMISELGLGYEKIHACP 218
++ L ++K +G SDKSFS LL ++ D PE N LPKS+Y+ KK++ + + Y KIHACP
Sbjct: 386 VLSLVNMKARYGCSDKSFSSLLQVVHDVLPEENTLPKSYYQAKKILCSMDMEY*KIHACP 565
Query: 219 NDCILYWDKYTNDEVCPKCGASRWKTVTEEVQPNGTKTSEKQKNTPLPKSVPLPGIPAKI 278
NDCILY ++ CP+CG SR+K +E + E K PL AK+
Sbjct: 566 NDCILYRHEFEEMSKCPRCGVSRYKVKDDE----ECSSDENSKKGPL----------AKV 703
Query: 279 LRWFPLKPRLQRLFMSSKIAEAMRWHRDG 307
L + P+ P+ +RLF + A+ + WH +G
Sbjct: 704 LWYLPIVPKFKRLFANGDDAKDLTWHANG 790
Score = 55.1 bits (131), Expect(2) = 7e-37
Identities = 38/132 (28%), Positives = 58/132 (43%), Gaps = 2/132 (1%)
Frame = +3
Query: 21 GVNEFLEFAFQNSQVSE--KIWCPCTTCANCKSLSRTSVYEHLTNPRRGFLRGYKQWIFH 78
GV +FL+ A + S+ E K +CPC C N + + +HL R Y WI+H
Sbjct: 12 GVEQFLQLASERSRPDENGKFFCPCINCLNGRRQKVNDIRDHLLCDE--IKRNYTTWIWH 185
Query: 79 GEKPVASSSTIEEPEHTEMEHDMDGLIHDVLASHTAEDPLFDDGERNPQVEENANKFYRL 138
GE S EP EM ++ +I D+ G+ + Q + +A + L
Sbjct: 186 GEMADMQSGPQSEPFDVEMRDRLEDMIRDL-------------GQESFQ-QAHAPMYDTL 323
Query: 139 VKDNEQILYPNC 150
D++ LYP C
Sbjct: 324 QIDSKTPLYPGC 359
>TC212179
Length = 764
Score = 62.8 bits (151), Expect = 8e-10
Identities = 38/95 (40%), Positives = 45/95 (47%), Gaps = 2/95 (2%)
Frame = +1
Query: 5 REWIRDPSRRSEEYIRGVNEFLEFAFQNSQVSE--KIWCPCTTCANCKSLSRTSVYEHLT 62
R W+ SR S EY GV +FL+FA Q Q E K +CPC C N + + EHL
Sbjct: 37 RSWMNQ-SRISPEYEEGVEQFLQFASQRGQPDEDGKYYCPCINCLNGRRQILDDIREHLL 213
Query: 63 NPRRGFLRGYKQWIFHGEKPVASSSTIEEPEHTEM 97
G R Y WI+HGE S EP EM
Sbjct: 214 CD--GIKRNYTTWIWHGEMTDMQSGQQSEPFDVEM 312
>BI425208
Length = 413
Score = 55.5 bits (132), Expect = 1e-07
Identities = 28/78 (35%), Positives = 42/78 (52%)
Frame = -3
Query: 3 ESREWIRDPSRRSEEYIRGVNEFLEFAFQNSQVSEKIWCPCTTCANCKSLSRTSVYEHLT 62
+++ WI+ + S EY++G+N+FLEF+F S+ I CPC C +R V L
Sbjct: 231 QNKGWIKLNIQASIEYLKGINDFLEFSFSYSKNKNTIRCPCCRCNAIYYKTRDEVIGDLL 52
Query: 63 NPRRGFLRGYKQWIFHGE 80
R F + Y+ W HGE
Sbjct: 51 KSR--FQQNYEHWESHGE 4
>TC227449 similar to UP|Q8S902 (Q8S902) Syringolide-induced protein 19-1-5,
partial (46%)
Length = 621
Score = 35.8 bits (81), Expect = 0.11
Identities = 17/51 (33%), Positives = 24/51 (46%)
Frame = -2
Query: 24 EFLEFAFQNSQVSEKIWCPCTTCANCKSLSRTSVYEHLTNPRRGFLRGYKQ 74
EF F + E WC C+ C +C S + SV L NP G R +++
Sbjct: 449 EFKMFGTLGEGLVEIFWCQCSNCKSCNSSAPNSVCPGLLNPTVGRTRHWRK 297
>TC213562 similar to UP|Q70US4 (Q70US4) NADH dehydrogenase 6, partial (18%)
Length = 546
Score = 33.9 bits (76), Expect = 0.42
Identities = 19/46 (41%), Positives = 27/46 (58%)
Frame = -1
Query: 840 STLDDGSFLHPMGRPLGRKRQQKFQLKKRKRVSRAKLEKKSVTTSS 885
+ LDDGS + R L RK+++K + K KRVS K +KK T +
Sbjct: 507 TALDDGSAIIISRRGLNRKKEKKNRTKN*KRVSNRKKKKKKNRTKN 370
>TC227448 homologue to UP|Q8S902 (Q8S902) Syringolide-induced protein 19-1-5,
partial (55%)
Length = 677
Score = 32.0 bits (71), Expect = 1.6
Identities = 14/38 (36%), Positives = 20/38 (51%)
Frame = -1
Query: 37 EKIWCPCTTCANCKSLSRTSVYEHLTNPRRGFLRGYKQ 74
E WC C+ C +C S + SV L NP G R +++
Sbjct: 662 EIYWCQCSNCKSCNSSAPNSVCLGLLNPTVG*TRHWRR 549
>TC219378 similar to UP|SLY1_ARATH (Q9SL48) SEC1-family transport protein
SLY1 (AtSLY1), partial (31%)
Length = 1203
Score = 31.2 bits (69), Expect = 2.7
Identities = 14/29 (48%), Positives = 18/29 (61%)
Frame = -3
Query: 257 SEKQKNTPLPKSVPLPGIPAKILRWFPLK 285
+EKQ+ P SVPLP +P WFP+K
Sbjct: 130 TEKQQYQPQTLSVPLPQLPLPTD*WFPMK 44
>TC225781 similar to GB|AAK11228.1|12963361|AF327524 copper-binding protein
CUTA {Arabidopsis thaliana;} , partial (63%)
Length = 1189
Score = 30.4 bits (67), Expect = 4.6
Identities = 18/40 (45%), Positives = 23/40 (57%)
Frame = +1
Query: 874 AKLEKKSVTTSSQICAF*L*PSHSICRVRHQSCFPSYTSS 913
AK +KK + TSS L PS CR+RH+ CFP + S
Sbjct: 706 AKKKKKKLETSS------LSPS---CRIRHEGCFP*WNQS 798
>BE823481
Length = 604
Score = 30.4 bits (67), Expect = 4.6
Identities = 15/49 (30%), Positives = 24/49 (48%)
Frame = +3
Query: 235 PKCGASRWKTVTEEVQPNGTKTSEKQKNTPLPKSVPLPGIPAKILRWFP 283
P+C S+ KT + QP +S + P PLP P+++L+ P
Sbjct: 105 PRCPCSQTKTHVDSXQPRVPISSTPKLFPSSPYPFPLPSTPSQVLQLSP 251
>TC215276 UP|143C_SOYBN (Q96452) 14-3-3-like protein C (SGF14C), complete
Length = 1379
Score = 30.0 bits (66), Expect = 6.1
Identities = 15/36 (41%), Positives = 20/36 (54%)
Frame = +3
Query: 253 GTKTSEKQKNTPLPKSVPLPGIPAKILRWFPLKPRL 288
GT T Q ++P+ + L IP K + W PL PRL
Sbjct: 654 GTITGTSQSSSPVMREKRLLIIP*KRISWLPLLPRL 761
>BI786841
Length = 422
Score = 29.6 bits (65), Expect = 7.9
Identities = 16/52 (30%), Positives = 28/52 (53%), Gaps = 7/52 (13%)
Frame = +1
Query: 45 TCANCKSLSRTSVYEHLTNPRRGFLRGYK-------QWIFHGEKPVASSSTI 89
+C CK+LS +YE ++N ++ FL +K I G+KP+ +T+
Sbjct: 241 SCLLCKTLSINLIYEPISNCKKCFLCYHKLRTYTTLHMIAVGKKPITQPTTV 396
>TC204092 homologue to UP|PSAD_NICSY (P29302) Photosystem I reaction center
subunit II, chloroplast precursor (Photosystem I 20 kDa
subunit) (PSI-D), partial (85%)
Length = 934
Score = 29.6 bits (65), Expect = 7.9
Identities = 14/43 (32%), Positives = 23/43 (52%)
Frame = -3
Query: 583 NYKARADLVDMGIRSILHPQPSPNTNTMLLPRACYQMTTKEKD 625
N R D V++ + +L P+PSP + +LLP ++ T D
Sbjct: 617 NLPIREDPVELVLDLVLGPEPSPKSQALLLPGQLEKVRTLPHD 489
>BU760638 weakly similar to GP|23237921|db contains ESTs D41667(S4320)
AU033165(S4320)~mucin-like protein, partial (27%)
Length = 430
Score = 29.6 bits (65), Expect = 7.9
Identities = 30/94 (31%), Positives = 44/94 (45%), Gaps = 1/94 (1%)
Frame = +3
Query: 587 RADLVDMGIRSILHPQPSPNTNTML-LPRACYQMTTKEKDDFLSVLKNVKAPDECSSNIS 645
R DLVD +I H ML LP A + + TK +L VK + I
Sbjct: 144 RQDLVDKFSSTIPHQMRLLLFLIMLHLPTALHSLRTK------IILWFVKLGNTGV*GIG 305
Query: 646 RCVQVQQRKIFGLKSYDCHVLMQELLPIALRGSL 679
Q + ++IF LK++ H+++ LLP+ GSL
Sbjct: 306 *KEQTKAQQIFSLKTFLVHLIISILLPMDPSGSL 407
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.336 0.147 0.497
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 63,146,726
Number of Sequences: 63676
Number of extensions: 1053133
Number of successful extensions: 9172
Number of sequences better than 10.0: 34
Number of HSP's better than 10.0 without gapping: 8982
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 9161
length of query: 1145
length of database: 12,639,632
effective HSP length: 108
effective length of query: 1037
effective length of database: 5,762,624
effective search space: 5975841088
effective search space used: 5975841088
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.7 bits)
S2: 64 (29.3 bits)
Lotus: description of TM0230.6


