

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0216.15
(500 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
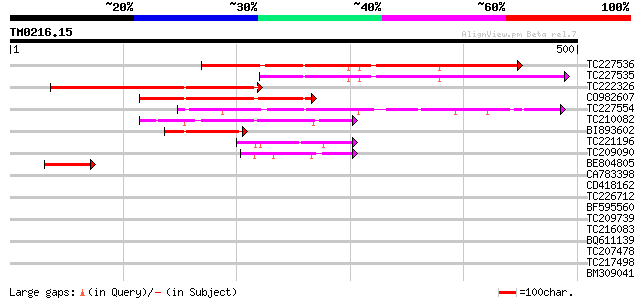
Score E
Sequences producing significant alignments: (bits) Value
TC227536 similar to UP|Q84RR0 (Q84RR0) ARIADNE-like protein ARI7... 252 2e-67
TC227535 similar to UP|Q84RR0 (Q84RR0) ARIADNE-like protein ARI7... 228 4e-60
TC222326 similar to UP|Q6RZU9 (Q6RZU9) ARIADNE-like protein, par... 183 1e-46
CO982607 175 4e-44
TC227554 similar to UP|Q84RR2 (Q84RR2) ARIADNE-like protein ARI2... 134 9e-32
TC210082 78 8e-15
BI893602 similar to GP|29125030|emb ARIADNE-like protein ARI8 {A... 67 2e-11
TC221196 weakly similar to UP|Q9FHR4 (Q9FHR4) Mutator-like trans... 54 1e-07
TC209090 52 6e-07
BE804805 weakly similar to GP|29125030|emb ARIADNE-like protein ... 45 1e-04
CA783398 41 0.001
CD418162 40 0.002
TC226712 34 0.17
BF595560 33 0.23
TC209739 similar to UP|Q8RVH7 (Q8RVH7) Aux/IAA protein, partial ... 32 0.51
TC216083 similar to UP|Q84P21 (Q84P21) 4-coumarate-CoA ligase-li... 30 1.9
BQ611139 homologue to GP|7862174|gb|A beta-ketoacyl-acyl carrier... 30 1.9
TC207478 similar to UP|Q6R567 (Q6R567) Ring domain containing pr... 30 1.9
TC217498 similar to UP|PEAM_SPIOL (Q9M571) Phosphoethanolamine N... 30 2.5
BM309041 similar to GP|14488069|gb| AT5g40270/MSN9_170 {Arabidop... 30 2.5
>TC227536 similar to UP|Q84RR0 (Q84RR0) ARIADNE-like protein ARI7, partial
(52%)
Length = 893
Score = 252 bits (644), Expect = 2e-67
Identities = 130/294 (44%), Positives = 179/294 (60%), Gaps = 11/294 (3%)
Frame = +3
Query: 170 VDRDMIRELASESSKVMYEQFLLWSYVDSKAKMRWCPAPGCDYVVCYQGDGGCARDLDVT 229
V +DMI LAS+ K Y+++LL SY++ K +WCPAPGC+Y V + G + DV+
Sbjct: 6 VGQDMINLLASDEDKEKYDRYLLRSYIEDNKKTKWCPAPGCEYAVTFDAGSG---NYDVS 176
Query: 230 CLCYHSFCWRCGEESHSPVDCETAGRWMKKIDKSTSENGKWILAHTKPCPRCKSLIEKNQ 289
CLC +SFCW C EE+H PVDC T +W+ K + + SEN WILA++KPCP+CK IEKNQ
Sbjct: 177 CLCSYSFCWNCTEEAHRPVDCGTVSKWILK-NSAESENMNWILANSKPCPKCKRPIEKNQ 353
Query: 290 GFKYMTCK--CGFQFCWVCF-----HEQLKCESRSCSPFIRMHIMEFQGT-DGEEVEINM 341
G +MTC C F+FCW+C H + +C+ R + +G D E M
Sbjct: 354 GCMHMTCTPPCKFEFCWLCLGAWSDHGERTGGFYACN---RYEAAKQEGVYDETERRREM 524
Query: 342 EKDNPKKCNHYYECWARNDFLSKSILQKLIDLDRKY---LSMSLQKRETYFDFVKKAWQQ 398
K++ ++ HYYE WA N + L L + + LS + + E+ F+ +AW Q
Sbjct: 525 AKNSLERYTHYYERWASNQSSRQKALADLHQMQTVHIEKLSDTQCQPESQLKFITEAWLQ 704
Query: 399 IAECRRILKWTNVYGYFLPKNEKAKKQFSEYIQVEAEVALEKLQQCAKVEETMF 452
I ECRR+LKWT YGY+LP++E AKKQF EY+Q EAE LE+L QCA+ E +F
Sbjct: 705 IVECRRVLKWTYSYGYYLPEHEHAKKQFFEYLQGEAESGLERLHQCAEKELQLF 866
>TC227535 similar to UP|Q84RR0 (Q84RR0) ARIADNE-like protein ARI7, partial
(54%)
Length = 1486
Score = 228 bits (582), Expect = 4e-60
Identities = 123/285 (43%), Positives = 167/285 (58%), Gaps = 12/285 (4%)
Frame = +2
Query: 221 GCARDLDVTCLCYHSFCWRCGEESHSPVDCETAGRWMKKIDKSTSENGKWILAHTKPCPR 280
G A + DV C C + FCW C EE+H PVDC T +W+ K + + SEN WILA++KPCP+
Sbjct: 5 GSAGNYDVACFCSYGFCWNCTEEAHRPVDCGTVAKWILK-NSAESENMNWILANSKPCPK 181
Query: 281 CKSLIEKNQGFKYMTCK--CGFQFCWVCF-----HEQLKCESRSCSPFIRMHIMEFQGT- 332
CK IEKNQG +MTC C F+FCW+C H + +C+ R + +G
Sbjct: 182 CKRPIEKNQGCMHMTCTPPCKFEFCWLCVGAWSDHGERTGGFYACN---RYEAAKQEGVY 352
Query: 333 DGEEVEINMEKDNPKKCNHYYECWARNDFLSKSILQKLIDLDRKY---LSMSLQKRETYF 389
D E M K++ ++ HYYE WA N + L L + + LS + E+
Sbjct: 353 DDTERRREMAKNSLERYTHYYERWASNQSSRQKALADLQQMQTVHIEKLSDIQCQPESQL 532
Query: 390 DFVKKAWQQIAECRRILKWTNVYGYFLPKNEKAKKQFSEYIQVEAEVALEKLQQCAKVE- 448
F+ +AW QI ECRR+LKWT YG++LP++E AKKQF EY+Q EAE LE+L QCA+ E
Sbjct: 533 KFITEAWLQIIECRRVLKWTYAYGFYLPEHEHAKKQFFEYLQGEAESGLERLHQCAEKEL 712
Query: 449 ETMFQDNYPEESLNKFQEKVIHLTDVTKIYFENLLRAFEDGLDKV 493
+ + P N F+ K+ LT VT+ YFENL+RA E+GL V
Sbjct: 713 QPFLSADDPSREFNDFRTKLAGLTSVTRNYFENLVRALENGLSDV 847
>TC222326 similar to UP|Q6RZU9 (Q6RZU9) ARIADNE-like protein, partial (36%)
Length = 557
Score = 183 bits (465), Expect = 1e-46
Identities = 88/188 (46%), Positives = 123/188 (64%), Gaps = 1/188 (0%)
Frame = +1
Query: 37 ENFTILSELEIKRLQEDAINKVASVLCIPDASACFLLCHNLWNVTKVLESWCEDEEKVQK 96
+NFTIL E +I+ QED + +VA+VL I A LL H+ W+V++V ++W DEE+V+K
Sbjct: 1 QNFTILKESDIRLRQEDDVTRVATVLSISRVFASILLRHHNWSVSRVHDTWFADEERVRK 180
Query: 97 AAGLLNQPKVEIDWLKDL-CQVCFRSSEKGKILSAGCAHPLCEACWMDYIDTNINKGPSK 155
A GLL +P V+ ++L C +CF + + +I A C HP C +CW YI T+IN GP
Sbjct: 181 AVGLLEKPIVQHPNTRELTCGICFENYPRARIEMASCGHPYCISCWEGYISTSINDGPG- 357
Query: 156 CLTLRCPDPSCDVAVDRDMIRELASESSKVMYEQFLLWSYVDSKAKMRWCPAPGCDYVVC 215
CL LRCPDP+CD A+ +DMI L S+ K Y ++LL SY++ K +WCPAPGC+Y V
Sbjct: 358 CLMLRCPDPTCDAAIGQDMINLLVSDEDKQKYARYLLRSYIEDNKKSKWCPAPGCEYAVT 537
Query: 216 YQGDGGCA 223
+ D G A
Sbjct: 538 F--DAGSA 555
>CO982607
Length = 842
Score = 175 bits (444), Expect = 4e-44
Identities = 75/156 (48%), Positives = 100/156 (64%)
Frame = -1
Query: 115 CQVCFRSSEKGKILSAGCAHPLCEACWMDYIDTNINKGPSKCLTLRCPDPSCDVAVDRDM 174
C +CF + + +I A C HP C +CW YI T+IN GP CL LRCPDP+CD A+ +DM
Sbjct: 749 CGICFENYPRARIEMASCGHPYCISCWEGYISTSINDGPG-CLMLRCPDPTCDAAIGQDM 573
Query: 175 IRELASESSKVMYEQFLLWSYVDSKAKMRWCPAPGCDYVVCYQGDGGCARDLDVTCLCYH 234
I L S+ K Y ++LL SY++ K +WCPAPGC+Y V + D G + DV+CLC +
Sbjct: 572 INLLVSDEDKQKYARYLLRSYIEDNKKSKWCPAPGCEYAVTF--DAGSTGNYDVSCLCSY 399
Query: 235 SFCWRCGEESHSPVDCETAGRWMKKIDKSTSENGKW 270
FCW C EE+H PVDC T +W+ K + + SEN W
Sbjct: 398 GFCWNCTEEAHRPVDCGTVAKWILK-NSAESENMNW 294
>TC227554 similar to UP|Q84RR2 (Q84RR2) ARIADNE-like protein ARI2, partial
(61%)
Length = 1678
Score = 134 bits (337), Expect = 9e-32
Identities = 96/365 (26%), Positives = 168/365 (45%), Gaps = 23/365 (6%)
Frame = +1
Query: 149 INKGPSKCLTLRCPDPSCDVAVDRDMIRELASESSKVM---YEQFLLWSYVDSKAKMRWC 205
IN+G SK +RC + C+ D ++R L S M YE+FLL SY++ +++WC
Sbjct: 16 INEGQSK--RIRCMEHKCNSICDEAVVRTLLSREHSHMAEKYERFLLESYIEDNKRVKWC 189
Query: 206 PA-PGCDYVVCYQGDGGCARDLDVTCLCYHSFCWRCGEESHSPVDCETAGRWMKKIDKST 264
P+ P C + + D C +V C C FC+ C E+HSP C W KK +
Sbjct: 190 PSTPHCGNAIRVEDDELC----EVECSCGVQFCFSCLSEAHSPCSCLMWELWAKKC-RDE 354
Query: 265 SENGKWILAHTKPCPRCKSLIEKNQGFKYMTCKCGFQFCWVC------FHEQLKCESRSC 318
SE WI HTKPCP+C +EKN G ++C CG FCW+C H SC
Sbjct: 355 SETVNWITVHTKPCPKCHKPVEKNGGCNLVSCICGQAFCWLCGGATGREHTWSSIAGHSC 534
Query: 319 SPFIRMHIMEFQGTDGEEVEINMEKDNPKKCNHYYECWARNDFLSKSILQKLIDLDRKYL 378
+ + E+ ++D + +++ A D S I KL + + +
Sbjct: 535 GRY----------KEQEKTAERAKRDLYRYMHYHNRYKAHTD--SFKIESKLKETIQGKI 678
Query: 379 SMSLQKRETYFDF--VKKAWQQIAECRRILKWTNVYGYFLPKNE-----------KAKKQ 425
++S +K T D+ V ++ RR+L ++ + +++ +E + K+
Sbjct: 679 AISEEKDSTLRDYSWVNNGLSRLFRSRRVLSYSYAFAFYMFGDELFKDEMTDAQREIKQN 858
Query: 426 FSEYIQVEAEVALEKLQQCAKVEETMFQDNYPEESLNKFQEKVIHLTDVTKIYFENLLRA 485
E Q + E +EKL +K+ E F + + ++ + + + ++++L+ + + +
Sbjct: 859 LFEDQQQQLEANVEKL---SKILEEPF-ETFSDDKVVEIRMQILNLSTIIDKLCQKMYEC 1026
Query: 486 FEDGL 490
E+ L
Sbjct: 1027IENDL 1041
>TC210082
Length = 779
Score = 78.2 bits (191), Expect = 8e-15
Identities = 59/197 (29%), Positives = 81/197 (40%), Gaps = 5/197 (2%)
Frame = +1
Query: 115 CQVCFRSSEKGKILSAGCAHPLCEACWMDYIDTNI-NKG--PSKCLTLRCPDPSCDVAVD 171
C +C E G L GC H C C ++ ++ I N+G P C C DP +
Sbjct: 43 CPICLCEVEDGYRLE-GCGHLFCRMCLVEQFESAIKNQGTFPVCCTHRDCGDP----ILL 207
Query: 172 RDMIRELASESSKVMYEQFLLWSYVDSKAKMRWCPAPGCDYVVCYQGDGGCARDLDVTCL 231
D+ L + + ++ L S R+CP+P C + D A + V
Sbjct: 208 TDLRSLLFGDKLEDLFRASLGAFVATSGGTYRFCPSPDCPSIYRV-ADPESAGEPFVCRA 384
Query: 232 CYHSFCWRCGEESHSPVDCETAGRWMKKIDKSTSE--NGKWILAHTKPCPRCKSLIEKNQ 289
CY C RC E H + CE + + D S E GK K C C +IEK
Sbjct: 385 CYSETCTRCHLEYHPYLSCERYKEFKEDPDSSLKEWCRGK---EQVKCCSACGYVIEKVD 555
Query: 290 GFKYMTCKCGFQFCWVC 306
G ++ CKCG CWVC
Sbjct: 556 GCNHVECKCGKHVCWVC 606
>BI893602 similar to GP|29125030|emb ARIADNE-like protein ARI8 {Arabidopsis
thaliana}, partial (10%)
Length = 423
Score = 66.6 bits (161), Expect = 2e-11
Identities = 33/73 (45%), Positives = 44/73 (60%)
Frame = -2
Query: 137 CEACWMDYIDTNINKGPSKCLTLRCPDPSCDVAVDRDMIRELASESSKVMYEQFLLWSYV 196
C + YI T+IN GP CL LRCPDP+C A+ +DMI L S+ K Y ++LL SY+
Sbjct: 236 CPFDMLGYISTSINDGPG-CLMLRCPDPTCGAAIGQDMINLLVSDEDKQKYARYLLRSYI 60
Query: 197 DSKAKMRWCPAPG 209
+ K+ CP G
Sbjct: 59 EDNKKV--CPRYG 27
>TC221196 weakly similar to UP|Q9FHR4 (Q9FHR4) Mutator-like transposase,
partial (20%)
Length = 758
Score = 54.3 bits (129), Expect = 1e-07
Identities = 35/117 (29%), Positives = 55/117 (46%), Gaps = 11/117 (9%)
Frame = +1
Query: 201 KMRWCPAPGCDYVVC------YQGD--GGCARDLDVTCL-CYHSFCWRCGEESHSPVDCE 251
K +CP P C ++ Y D G + CL C FC+ C HS + C
Sbjct: 127 KKIYCPYPTCSALMSKTEVLEYSKDITGQSEQSEPKICLKCRGLFCFNCKVPWHSGMTCN 306
Query: 252 TAGRWMKKIDKSTSENGKWILAHT--KPCPRCKSLIEKNQGFKYMTCKCGFQFCWVC 306
T R M I + K++ + + + C +C +IE +G +MTC+CG++FC+ C
Sbjct: 307 TYKR-MNPIPPAEDLKLKFLASRSLWQQCLKCNHMIELAEGCYHMTCRCGYEFCYNC 474
>TC209090
Length = 1011
Score = 52.0 bits (123), Expect = 6e-07
Identities = 34/118 (28%), Positives = 50/118 (41%), Gaps = 15/118 (12%)
Frame = +1
Query: 204 WCPAPGCDYVV----CYQGDGGCARDLDVTCL----CYHSFCWRCGEESHSPVDC-ETAG 254
+CP P C ++ C + D +C+ C C C HS + C E
Sbjct: 55 YCPFPNCSVLLDPHECSSARASSSSQSDNSCIECPVCRRFICVDCKVPWHSSMSCLEYQN 234
Query: 255 RWMKKIDKST------SENGKWILAHTKPCPRCKSLIEKNQGFKYMTCKCGFQFCWVC 306
K+ D S ++N +W K C +C+ IE QG +MTC CG +FC+ C
Sbjct: 235 LPEKERDVSDITLHRLAQNKRW-----KRCQQCRRTIELTQGCYHMTCWCGHEFCYSC 393
Score = 28.5 bits (62), Expect = 7.3
Identities = 9/16 (56%), Positives = 11/16 (68%)
Frame = +1
Query: 228 VTCLCYHSFCWRCGEE 243
+TC C H FC+ CG E
Sbjct: 355 MTCWCGHEFCYSCGAE 402
>BE804805 weakly similar to GP|29125030|emb ARIADNE-like protein ARI8
{Arabidopsis thaliana}, partial (10%)
Length = 328
Score = 44.7 bits (104), Expect = 1e-04
Identities = 23/45 (51%), Positives = 31/45 (68%)
Frame = +2
Query: 31 EEEDKRENFTILSELEIKRLQEDAINKVASVLCIPDASACFLLCH 75
E +NFTIL+EL+I++ Q+D I +VA+VL IP SA LL H
Sbjct: 158 ESRRPEQNFTILTELDIRQRQDDDIARVAAVLSIPWVSASILLRH 292
>CA783398
Length = 457
Score = 41.2 bits (95), Expect = 0.001
Identities = 26/128 (20%), Positives = 56/128 (43%), Gaps = 1/128 (0%)
Frame = +3
Query: 114 LCQVCFRSSEKGKILSA-GCAHPLCEACWMDYIDTNINKGPSKCLTLRCPDPSCDVAVDR 172
+C++C + S GC H C +C Y+++ + + +++ CP P C ++
Sbjct: 87 VCEICTETKTARDSFSIIGCHHVYCNSCVAQYVESKLEEN---IVSIPCPVPGCRGLLEA 257
Query: 173 DMIRELASESSKVMYEQFLLWSYVDSKAKMRWCPAPGCDYVVCYQGDGGCARDLDVTCLC 232
D RE+ + + + L + + ++ K +CP C ++ + R+ + C
Sbjct: 258 DDCREILAPRVFDRWGKVLCEAVIAAEEKF-YCPFADCSVMLIRGIEENNIREAECP-NC 431
Query: 233 YHSFCWRC 240
FC +C
Sbjct: 432 RRLFCAQC 455
>CD418162
Length = 364
Score = 40.0 bits (92), Expect = 0.002
Identities = 19/55 (34%), Positives = 26/55 (46%), Gaps = 2/55 (3%)
Frame = -1
Query: 254 GRWMKKIDKSTSENGKWILA--HTKPCPRCKSLIEKNQGFKYMTCKCGFQFCWVC 306
GR+ + + S +W K C C +IEK G ++ CKCG CWVC
Sbjct: 358 GRYQEFKEDPDSSLKEWCRGKEQVKCCSACGYVIEKVDGCNHVECKCGKHVCWVC 194
>TC226712
Length = 1402
Score = 33.9 bits (76), Expect = 0.17
Identities = 12/32 (37%), Positives = 16/32 (49%), Gaps = 3/32 (9%)
Frame = +3
Query: 278 CPRCKSLIEKNQGFKYMT---CKCGFQFCWVC 306
C C+S I KN Y C+C +CW+C
Sbjct: 66 CIHCQSFIRKNMDHAYYLS*ICRCSCHYCWIC 161
>BF595560
Length = 422
Score = 33.5 bits (75), Expect = 0.23
Identities = 21/85 (24%), Positives = 36/85 (41%), Gaps = 1/85 (1%)
Frame = +2
Query: 159 LRCPDPSCDVAVDRDMIRELASESSKVMYEQFLLWSYVDSKAKMRWCPAPGCDYVVCYQG 218
L+CP+ C + +++ ++ +E +L + S + +CP C+ C G
Sbjct: 185 LQCPEAKCACMIPTGLLKPFLDDTDYERWESMILEKTLASMSDAVYCPR--CE-TPCIDG 355
Query: 219 DGGCARDLDVTC-LCYHSFCWRCGE 242
+ D C CY SFC C E
Sbjct: 356 E-----DQHAQCPKCYFSFCTLCRE 415
>TC209739 similar to UP|Q8RVH7 (Q8RVH7) Aux/IAA protein, partial (55%)
Length = 953
Score = 32.3 bits (72), Expect = 0.51
Identities = 17/63 (26%), Positives = 33/63 (51%), Gaps = 2/63 (3%)
Frame = -3
Query: 57 KVASVLCIPDASACFLLC--HNLWNVTKVLESWCEDEEKVQKAAGLLNQPKVEIDWLKDL 114
++ S+ C P +C+ C H+L +++ ++C ++ V A G L K ++W K
Sbjct: 333 RLESLPCTPSQRSCYDRCCNHSLQQKQQLVNNYCSQKDGVVVAIGPLK*KKQHVEWQK-- 160
Query: 115 CQV 117
C+V
Sbjct: 159 CRV 151
>TC216083 similar to UP|Q84P21 (Q84P21) 4-coumarate-CoA ligase-like protein,
partial (80%)
Length = 1411
Score = 30.4 bits (67), Expect = 1.9
Identities = 20/78 (25%), Positives = 31/78 (39%), Gaps = 3/78 (3%)
Frame = +3
Query: 232 CYHSFCWRCGEESHSPVDCETAGRWMKKIDKSTSENGKWILAHTKPCP---RCKSLIEKN 288
C+ WR + S++ C T W S S G W+ A C RC + ++
Sbjct: 666 CWVGSTWRRTKPSYARSPCFTYTAW------SPSPRGFWLPAPPSSCSPSSRCTTCSPRS 827
Query: 289 QGFKYMTCKCGFQFCWVC 306
+G +T + F W C
Sbjct: 828 RGSAPLTSRSCRPFWWPC 881
>BQ611139 homologue to GP|7862174|gb|A beta-ketoacyl-acyl carrier protein
synthase III {Glycine max}, partial (14%)
Length = 420
Score = 30.4 bits (67), Expect = 1.9
Identities = 15/54 (27%), Positives = 26/54 (47%), Gaps = 5/54 (9%)
Frame = +2
Query: 346 PKKCNHYYECWARNDFLSKSI-----LQKLIDLDRKYLSMSLQKRETYFDFVKK 394
P CN + CW+ F + S+ L ++ DL++ L + L K T+ +K
Sbjct: 119 PDYCNGWLRCWSYLGFSNSSLGLNAFLSRINDLEKNQLQVQLSKYPTFLHNCRK 280
>TC207478 similar to UP|Q6R567 (Q6R567) Ring domain containing protein,
partial (51%)
Length = 1103
Score = 30.4 bits (67), Expect = 1.9
Identities = 23/80 (28%), Positives = 36/80 (44%)
Frame = +1
Query: 209 GCDYVVCYQGDGGCARDLDVTCLCYHSFCWRCGEESHSPVDCETAGRWMKKIDKSTSENG 268
G D +C + C +D VT LC H +CW C +W+ + ++SEN
Sbjct: 238 GFDCNICLE----CVQDPVVT-LCGHLYCWPC------------IYKWL-NLQTASSENE 363
Query: 269 KWILAHTKPCPRCKSLIEKN 288
+ + CP CKS I ++
Sbjct: 364 E----EKQQCPVCKSEISQS 411
>TC217498 similar to UP|PEAM_SPIOL (Q9M571) Phosphoethanolamine
N-methyltransferase , partial (65%)
Length = 1509
Score = 30.0 bits (66), Expect = 2.5
Identities = 12/30 (40%), Positives = 16/30 (53%)
Frame = -1
Query: 187 YEQFLLWSYVDSKAKMRWCPAPGCDYVVCY 216
Y + LLW Y + +K RW P CD C+
Sbjct: 249 YRKILLWLY-STLSKKRWKPLSSCDLTFCH 163
>BM309041 similar to GP|14488069|gb| AT5g40270/MSN9_170 {Arabidopsis
thaliana}, partial (15%)
Length = 416
Score = 30.0 bits (66), Expect = 2.5
Identities = 14/30 (46%), Positives = 19/30 (62%), Gaps = 3/30 (10%)
Frame = +3
Query: 61 VLCIPDAS---ACFLLCHNLWNVTKVLESW 87
VLCIPD S AC L N+W K++++W
Sbjct: 285 VLCIPDLSILLACIGLPVNVWKSLKLIKAW 374
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.321 0.136 0.439
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 29,231,970
Number of Sequences: 63676
Number of extensions: 529927
Number of successful extensions: 3196
Number of sequences better than 10.0: 59
Number of HSP's better than 10.0 without gapping: 3068
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3166
length of query: 500
length of database: 12,639,632
effective HSP length: 101
effective length of query: 399
effective length of database: 6,208,356
effective search space: 2477134044
effective search space used: 2477134044
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0216.15


