

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
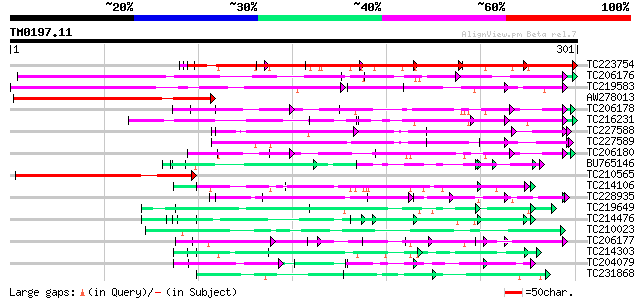
Query= TM0197.11
(301 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC223754 similar to UP|Q86EQ4 (Q86EQ4) Clone ZZD1536 mRNA sequen... 245 2e-65
TC206176 weakly similar to UP|Q8LPA7 (Q8LPA7) Cold shock protein... 209 2e-54
TC219583 weakly similar to UP|GRP2_NICSY (P27484) Glycine-rich p... 162 2e-40
AW278013 similar to SP|P27484|GRP2 Glycine-rich protein 2. [Wood... 111 3e-25
TC206178 similar to UP|Q8LF59 (Q8LF59) DNA-binding protein, part... 108 2e-24
TC216231 similar to UP|Q41188 (Q41188) Glycine-rich protein 2 (G... 108 3e-24
TC227588 similar to PIR|T00837|T00837 glycine-rich protein T13L1... 100 1e-21
TC227589 94 6e-20
TC206180 similar to UP|Q8LF59 (Q8LF59) DNA-binding protein, part... 91 6e-19
BU765146 similar to GP|21322711|e pherophorin-dz1 protein {Volvo... 84 8e-17
TC210565 similar to GB|AAO27142.1|27904309|AE014017 cold shock p... 84 8e-17
TC214106 weakly similar to UP|Q8L685 (Q8L685) Pherophorin-dz1 pr... 81 7e-16
TC228935 similar to UP|Q9FHC2 (Q9FHC2) Arabidopsis thaliana geno... 74 6e-14
TC219649 weakly similar to UP|Q95JD1 (Q95JD1) Basic proline-rich... 74 1e-13
TC214476 weakly similar to UP|Q8L685 (Q8L685) Pherophorin-dz1 pr... 70 2e-12
TC210023 weakly similar to UP|Q868B4 (Q868B4) C. elegans GRL-25 ... 63 1e-10
TC206177 similar to UP|Q41188 (Q41188) Glycine-rich protein 2 (G... 62 3e-10
TC214303 similar to UP|Q9T0K5 (Q9T0K5) Extensin-like protein, pa... 61 5e-10
TC204079 similar to UP|Q9SWA8 (Q9SWA8) Glycine-rich RNA-binding ... 59 3e-09
TC231868 similar to UP|Q93424 (Q93424) C. elegans GRL-23 protein... 55 5e-08
>TC223754 similar to UP|Q86EQ4 (Q86EQ4) Clone ZZD1536 mRNA sequence, partial
(22%)
Length = 742
Score = 245 bits (625), Expect = 2e-65
Identities = 112/152 (73%), Positives = 121/152 (78%), Gaps = 8/152 (5%)
Frame = +2
Query: 158 SGGGGAGCYNCGDTGHLARDCNRSNNSGGGGGG-GGACYTCGGFGHLARDCMRGGNGG-- 214
+GGGGA CY CG+ GHLARDCNRS+NSGGGGGG GG C+ CGGFGHLARDC+RGG G
Sbjct: 23 NGGGGAACYQCGEFGHLARDCNRSSNSGGGGGGSGGGCFNCGGFGHLARDCVRGGGGSVG 202
Query: 215 -GGGPGSASCFRCGGIGHMARDCATAKGPSSGGVGGGGCFRCGEVGHLARDC--DGGVAV 271
GGG G SCFRCGG GHMARDCAT KG GG GGGCFRCGEVGHLARDC +GG
Sbjct: 203 IGGGGGGGSCFRCGGFGHMARDCATGKGNIGGGGSGGGCFRCGEVGHLARDCGMEGGRFG 382
Query: 272 SGG--GGNAGRNTCFNCGKPGHFARECIEASG 301
GG GG G++TCFNCGKPGHFAREC+EASG
Sbjct: 383 GGGGSGGGGGKSTCFNCGKPGHFARECVEASG 478
Score = 193 bits (490), Expect = 9e-50
Identities = 94/157 (59%), Positives = 105/157 (66%), Gaps = 13/157 (8%)
Frame = +2
Query: 132 GGGGAACYNCGDAGHLARDCNRSNNNSGGGGA---GCYNCGDTGHLARDCNRSNNSG--- 185
GGGGAACY CG+ GHLARDCNRS+N+ GGGG GC+NCG GHLARDC R
Sbjct: 26 GGGGAACYQCGEFGHLARDCNRSSNSGGGGGGSGGGCFNCGGFGHLARDCVRGGGGSVGI 205
Query: 186 GGGGGGGACYTCGGFGHLARDCMRG-GNGGGGGPGSASCFRCGGIGHMARDCATAKGPSS 244
GGGGGGG+C+ CGGFGH+ARDC G GN GGGG G CFRCG +GH+ARDC G
Sbjct: 206 GGGGGGGSCFRCGGFGHMARDCATGKGNIGGGGSG-GGCFRCGEVGHLARDCGMEGGRFG 382
Query: 245 GGVGGGG------CFRCGEVGHLARDCDGGVAVSGGG 275
GG G GG CF CG+ GH AR+C V SG G
Sbjct: 383 GGGGSGGGGGKSTCFNCGKPGHFAREC---VEASG*G 484
Score = 189 bits (481), Expect = 1e-48
Identities = 88/160 (55%), Positives = 101/160 (63%), Gaps = 13/160 (8%)
Frame = +2
Query: 95 GERRNGGGGGGGGGGCYNCGDTGHLARDCHRSNNNGGGGGGA--ACYNCGDAGHLARDCN 152
G RRNGGGG CY CG+ GHLARDC+RS+N+GGGGGG+ C+NCG GHLARDC
Sbjct: 11 GARRNGGGGAA----CYQCGEFGHLARDCNRSSNSGGGGGGSGGGCFNCGGFGHLARDCV 178
Query: 153 RSNNNS-----GGGGAGCYNCGDTGHLARDCNRSNNSGGGGGGGGACYTCGGFGHLARDC 207
R S GGGG C+ CG GH+ARDC + GGGG GG C+ CG GHLARDC
Sbjct: 179 RGGGGSVGIGGGGGGGSCFRCGGFGHMARDCATGKGNIGGGGSGGGCFRCGEVGHLARDC 358
Query: 208 ------MRGGNGGGGGPGSASCFRCGGIGHMARDCATAKG 241
GG G GGG G ++CF CG GH AR+C A G
Sbjct: 359 GMEGGRFGGGGGSGGGGGKSTCFNCGKPGHFARECVEASG 478
Score = 151 bits (382), Expect = 3e-37
Identities = 70/131 (53%), Positives = 81/131 (61%), Gaps = 12/131 (9%)
Frame = +2
Query: 99 NGGGGGGGGGGCYNCGDTGHLARDCHRSNNN----GGGGGGAACYNCGDAGHLARDCNRS 154
+GGGGGG GGGC+NCG GHLARDC R GGGGGG +C+ CG GH+ARDC
Sbjct: 101 SGGGGGGSGGGCFNCGGFGHLARDCVRGGGGSVGIGGGGGGGSCFRCGGFGHMARDCATG 280
Query: 155 NNNSGGGGAG--CYNCGDTGHLARDCN----RSNNSGGGGGGGG--ACYTCGGFGHLARD 206
N GGGG+G C+ CG+ GHLARDC R GG GGGGG C+ CG GH AR+
Sbjct: 281 KGNIGGGGSGGGCFRCGEVGHLARDCGMEGGRFGGGGGSGGGGGKSTCFNCGKPGHFARE 460
Query: 207 CMRGGNGGGGG 217
C+ G G
Sbjct: 461 CVEASG*GEEG 493
Score = 112 bits (279), Expect = 3e-25
Identities = 51/105 (48%), Positives = 59/105 (55%), Gaps = 9/105 (8%)
Frame = +2
Query: 93 RGGERRNGGGGGGGGGGCYNCGDTGHLARDCHRSNNN-GGGGGGAACYNCGDAGHLARDC 151
RGG G GGGGGGG C+ CG GH+ARDC N GGGG G C+ CG+ GHLARDC
Sbjct: 179 RGGGGSVGIGGGGGGGSCFRCGGFGHMARDCATGKGNIGGGGSGGGCFRCGEVGHLARDC 358
Query: 152 NRSNNNSGGGGAG--------CYNCGDTGHLARDCNRSNNSGGGG 188
GGGG C+NCG GH AR+C ++ G G
Sbjct: 359 GMEGGRFGGGGGSGGGGGKSTCFNCGKPGHFARECVEASG*GEEG 493
Score = 72.0 bits (175), Expect = 3e-13
Identities = 31/61 (50%), Positives = 37/61 (59%)
Frame = +2
Query: 241 GPSSGGVGGGGCFRCGEVGHLARDCDGGVAVSGGGGNAGRNTCFNCGKPGHFARECIEAS 300
G G GG C++CGE GHLARDC+ GGGG +G CFNCG GH AR+C+
Sbjct: 11 GARRNGGGGAACYQCGEFGHLARDCNRSSNSGGGGGGSG-GGCFNCGGFGHLARDCVRGG 187
Query: 301 G 301
G
Sbjct: 188 G 190
Score = 49.3 bits (116), Expect = 2e-06
Identities = 25/51 (49%), Positives = 29/51 (56%), Gaps = 3/51 (5%)
Frame = +2
Query: 91 GWRGGERRNGGGGGGGGGG---CYNCGDTGHLARDCHRSNNNGGGGGGAAC 138
G GG R GGGG GGGGG C+NCG GH AR+C ++ G G C
Sbjct: 359 GMEGG-RFGGGGGSGGGGGKSTCFNCGKPGHFARECVEASG*GEEGFVTFC 508
>TC206176 weakly similar to UP|Q8LPA7 (Q8LPA7) Cold shock protein-1, partial
(77%)
Length = 883
Score = 209 bits (531), Expect = 2e-54
Identities = 118/238 (49%), Positives = 138/238 (57%), Gaps = 3/238 (1%)
Frame = +3
Query: 5 RFSGVVQWFNNVKGFGFIKPDDGGEDLFVHQSSIRSDGYRTLLEGDLVEFSIATGDNDKT 64
R SG V+WFN+ KGFGFI PDDG +DLFVHQS I+SDG+R+L EG+ VEF+I + + +
Sbjct: 57 RVSGKVKWFNDQKGFGFISPDDGSDDLFVHQSQIKSDGFRSLAEGESVEFAIESESDGRA 236
Query: 65 KAVDVTGPNGAALQPTRKDSAPRGFGGWRGGERRNGGGGGGGGGGCYNCGDTGHLARDCH 124
KAVDVTGP+GA++Q TR+ G GG G R GGGGG GGGG Y
Sbjct: 237 KAVDVTGPDGASVQGTRRG----GDGGRSYGGGRGGGGGGYGGGGGYG------------ 368
Query: 125 RSNNNGGGGGGAACYNCGDAGHLARDCNRSNNNSGGGGAGCYNCGDTGHLARDCNRSNNS 184
GGG GG Y G GGGG CYNCG++GHLARDC S
Sbjct: 369 ----GGGGYGGGGGYGGG--------------GRGGGGGACYNCGESGHLARDC--SGGG 488
Query: 185 GG---GGGGGGACYTCGGFGHLARDCMRGGNGGGGGPGSASCFRCGGIGHMARDCATA 239
GG GGGGGG Y GG G GG GGGGG G SC+ CG GH ARDC ++
Sbjct: 489 GGDRYGGGGGGGRYGGGGGGRYV-----GGGGGGGGGG--SCYSCGESGHFARDCPSS 641
Score = 89.0 bits (219), Expect = 2e-18
Identities = 54/131 (41%), Positives = 65/131 (49%), Gaps = 11/131 (8%)
Frame = +3
Query: 177 DCNRSNNSGGGGGGGGACYTCGGFGHLARDCMRGGNGGGG----------GPGSASCFRC 226
D RS G GGGGGG GG G+ GG GG G G G +C+ C
Sbjct: 297 DGGRSYGGGRGGGGGGYG---GGGGY-------GGGGGYGGGGGYGGGGRGGGGGACYNC 446
Query: 227 GGIGHMARDCATAKGPSS-GGVGGGGCFRCGEVGHLARDCDGGVAVSGGGGNAGRNTCFN 285
G GH+ARDC+ G GG GGGG + G GG V GGGG G +C++
Sbjct: 447 GESGHLARDCSGGGGGDRYGGGGGGGRYGGG---------GGGRYVGGGGGGGGGGSCYS 599
Query: 286 CGKPGHFAREC 296
CG+ GHFAR+C
Sbjct: 600 CGESGHFARDC 632
Score = 53.5 bits (127), Expect = 1e-07
Identities = 38/116 (32%), Positives = 44/116 (37%), Gaps = 3/116 (2%)
Frame = +3
Query: 189 GGGGACYTCGGFGHLARDCMRGGNGG---GGGPGSASCFRCGGIGHMARDCATAKGPSSG 245
G A G G + RGG+GG GGG G GG G G
Sbjct: 228 GRAKAVDVTGPDGASVQGTRRGGDGGRSYGGGRGGGG----GGYG------------GGG 359
Query: 246 GVGGGGCFRCGEVGHLARDCDGGVAVSGGGGNAGRNTCFNCGKPGHFARECIEASG 301
G GGGG + GG GGG G C+NCG+ GH AR+C G
Sbjct: 360 GYGGGGGY------------GGGGGYGGGGRGGGGGACYNCGESGHLARDCSGGGG 491
>TC219583 weakly similar to UP|GRP2_NICSY (P27484) Glycine-rich protein 2,
partial (45%)
Length = 995
Score = 162 bits (410), Expect = 2e-40
Identities = 93/192 (48%), Positives = 107/192 (55%), Gaps = 14/192 (7%)
Frame = +2
Query: 1 MAEERFS-GVVQWFNNVKGFGFIKPDDGGEDLFVHQSSIRSDGYRTLLEGDLVEFSIATG 59
MAE R S G V+WFN KGFGFI P DG +DLFVH +SIRSDGYR+L +G VEF + G
Sbjct: 83 MAETRRSTGTVKWFNAHKGFGFITPQDGTDDLFVHFTSIRSDGYRSLSDGQSVEFLLDYG 262
Query: 60 DNDKTKAVDVTGPNGAALQPTRKDSAPRGFGGWRGGERRNGGGGGGGGGGCYNCGDTGHL 119
D+ +T AVDVT + L GG+RGG GGG G GGG Y G+
Sbjct: 263 DDGRTMAVDVTSAVRSRLP-----------GGFRGG-----GGGRGRGGGRYGGGEG--- 385
Query: 120 ARDCHRSNNNGGGGGGAACYNCGDAGHLARDC-------------NRSNNNSGGGGAGCY 166
R G GGG CYNCG GHLARDC NR GGGG GC+
Sbjct: 386 -----RGRGFGRRGGGPECYNCGRIGHLARDCYHGQGGGGGDDGRNRRRGGGGGGGGGCF 550
Query: 167 NCGDTGHLARDC 178
NCG+ GH AR+C
Sbjct: 551 NCGEEGHFAREC 586
Score = 75.1 bits (183), Expect = 4e-14
Identities = 45/99 (45%), Positives = 50/99 (50%), Gaps = 12/99 (12%)
Frame = +2
Query: 210 GGNGGGGGPGSASCFRCGGIGHMARDCATAKGPSSGGVGGGG-CFRCGEVGHLARDCDGG 268
G GGGGG G GG G +G G GGG C+ CG +GHLARDC G
Sbjct: 323 GFRGGGGGRGRGGGRYGGGEG---------RGRGFGRRGGGPECYNCGRIGHLARDCYHG 475
Query: 269 VAVSGGGGNAGRNT-----------CFNCGKPGHFAREC 296
GGGG+ GRN CFNCG+ GHFAREC
Sbjct: 476 QG--GGGGDDGRNRRRGGGGGGGGGCFNCGEEGHFAREC 586
Score = 74.7 bits (182), Expect = 5e-14
Identities = 43/97 (44%), Positives = 47/97 (48%), Gaps = 11/97 (11%)
Frame = +2
Query: 180 RSNNSGGGGGGGGACYTCGGFGHLARDCMRGGNGGGGGPGSASCFRCGGIGHMARDCATA 239
RS GG GGGG GG RG GGGP C+ CG IGH+ARDC
Sbjct: 305 RSRLPGGFRGGGGGRGRGGGRYGGGEGRGRGFGRRGGGP---ECYNCGRIGHLARDCYHG 475
Query: 240 KGPSSG-----------GVGGGGCFRCGEVGHLARDC 265
+G G G GGGGCF CGE GH AR+C
Sbjct: 476 QGGGGGDDGRNRRRGGGGGGGGGCFNCGEEGHFAREC 586
Score = 37.0 bits (84), Expect = 0.011
Identities = 27/71 (38%), Positives = 32/71 (45%), Gaps = 1/71 (1%)
Frame = +2
Query: 232 MARDCATA-KGPSSGGVGGGGCFRCGEVGHLARDCDGGVAVSGGGGNAGRNTCFNCGKPG 290
MA D +A + GG GGG R G G R G G G G C+NCG+ G
Sbjct: 278 MAVDVTSAVRSRLPGGFRGGGGGR-GRGG--GRYGGGEGRGRGFGRRGGGPECYNCGRIG 448
Query: 291 HFARECIEASG 301
H AR+C G
Sbjct: 449 HLARDCYHGQG 481
>AW278013 similar to SP|P27484|GRP2 Glycine-rich protein 2. [Wood tobacco]
{Nicotiana sylvestris}, partial (48%)
Length = 344
Score = 111 bits (278), Expect = 3e-25
Identities = 58/107 (54%), Positives = 73/107 (68%)
Frame = +1
Query: 3 EERFSGVVQWFNNVKGFGFIKPDDGGEDLFVHQSSIRSDGYRTLLEGDLVEFSIATGDND 62
++R +G V+WF++ KGFGFI DD GE+ FVHQSSIRSDG+R+L G+ VEF I +
Sbjct: 28 KQRVTGKVKWFDDQKGFGFITTDDSGEEFFVHQSSIRSDGFRSLALGESVEFLIDSDPEG 207
Query: 63 KTKAVDVTGPNGAALQPTRKDSAPRGFGGWRGGERRNGGGGGGGGGG 109
+TKAVDVTGP+ + +Q TR G GG RG GG GG GGGG
Sbjct: 208 RTKAVDVTGPDESPVQGTR-----GGGGGGRGSYGGGGGRGGYGGGG 333
Score = 30.0 bits (66), Expect = 1.3
Identities = 17/34 (50%), Positives = 17/34 (50%)
Frame = +1
Query: 186 GGGGGGGACYTCGGFGHLARDCMRGGNGGGGGPG 219
GGGGGG Y GG RGG GGGG G
Sbjct: 265 GGGGGGRGSYGGGG--------GRGGYGGGGRGG 342
>TC206178 similar to UP|Q8LF59 (Q8LF59) DNA-binding protein, partial (69%)
Length = 1138
Score = 108 bits (271), Expect = 2e-24
Identities = 70/203 (34%), Positives = 85/203 (41%), Gaps = 16/203 (7%)
Frame = +2
Query: 110 CYNCGDTGHLARDCHRSNNNGGGGGGAACYNCGDAGHLARDCNRSNNNSGGGGAGCYNCG 169
C+NC + GH+A C N G C+ CG AGH AR+C+ G C NC
Sbjct: 458 CWNCKEPGHMASSCP---NEG------ICHTCGKAGHRARECSAPPMPPGDLRL-CNNCY 607
Query: 170 DTGHLARDCNRSNNSGGGGGGGGACYTCGGFGHLARDCMRGGNGGGGGPGSASCFRCGGI 229
GH+A +C AC C GHLARDC P C C
Sbjct: 608 KQGHIAAECTNEK----------ACNNCRKTGHLARDC----------PNDPICNLCNVS 727
Query: 230 GHMARDCATAK--GPSSGGVGGGG--------------CFRCGEVGHLARDCDGGVAVSG 273
GH+AR C A G SGG GGGG C C ++GH++RDC G + +
Sbjct: 728 GHVARQCPKANVLGDRSGGGGGGGGARGGGGGGYRDVVCRNCQQLGHMSRDCMGPLMI-- 901
Query: 274 GGGNAGRNTCFNCGKPGHFAREC 296
C NCG GH A EC
Sbjct: 902 ---------CHNCGGRGHLAYEC 943
Score = 94.0 bits (232), Expect = 8e-20
Identities = 62/163 (38%), Positives = 71/163 (43%), Gaps = 4/163 (2%)
Frame = +2
Query: 110 CYNCGDTGHLARDCHRSNNNGGGGGGAACYNCGDAGHLARDCNRSNNNSGGGGAGCYNCG 169
C NC GH+A +C AC NC GHLARDC C C
Sbjct: 593 CNNCYKQGHIAAECTNEK---------ACNNCRKTGHLARDCPNDPI--------CNLCN 721
Query: 170 DTGHLARDCNRSN----NSGGGGGGGGACYTCGGFGHLARDCMRGGNGGGGGPGSASCFR 225
+GH+AR C ++N SGGGGGGGGA RGG GGGG C
Sbjct: 722 VSGHVARQCPKANVLGDRSGGGGGGGGA---------------RGG--GGGGYRDVVCRN 850
Query: 226 CGGIGHMARDCATAKGPSSGGVGGGGCFRCGEVGHLARDCDGG 268
C +GHM+RDC GP C CG GHLA +C G
Sbjct: 851 CQQLGHMSRDC---MGPLM------ICHNCGGRGHLAYECPSG 952
Score = 92.8 bits (229), Expect = 2e-19
Identities = 63/205 (30%), Positives = 81/205 (38%), Gaps = 1/205 (0%)
Frame = +2
Query: 97 RRNGGGGGGGGGGCYNCGDTGHLARDCHRSNNNGGGGGGAACYNCGDAGHLARDCNRSNN 156
RR+ G C NC GH AR+C A C+NCG GH+A +C +
Sbjct: 305 RRDSRRGFSRDNLCKNCKRPGHYARECPNV---------AICHNCGLPGHIASECTTKSL 457
Query: 157 NSGGGGAGCYNCGDTGHLARDCNRSNNSGGGGGGGGACYTCGGFGHLARDCMRGGNGGGG 216
C+NC + GH+A C G C+TCG GH AR+C +
Sbjct: 458 --------CWNCKEPGHMASSCPNE----------GICHTCGKAGHRAREC----SAPPM 571
Query: 217 GPGSAS-CFRCGGIGHMARDCATAKGPSSGGVGGGGCFRCGEVGHLARDCDGGVAVSGGG 275
PG C C GH+A +C K C C + GHLARDC
Sbjct: 572 PPGDLRLCNNCYKQGHIAAECTNEKA----------CNNCRKTGHLARDCPNDPI----- 706
Query: 276 GNAGRNTCFNCGKPGHFARECIEAS 300
C C GH AR+C +A+
Sbjct: 707 -------CNLCNVSGHVARQCPKAN 760
Score = 61.6 bits (148), Expect = 4e-10
Identities = 43/130 (33%), Positives = 50/130 (38%), Gaps = 9/130 (6%)
Frame = +2
Query: 176 RDCNRSNNSGGGGGGGGACYTCGGFGHLARDCMRGGNGGGGGPGSASCFRCGGIGHMARD 235
RD +S G C C GH AR+C P A C CG GH+A +
Sbjct: 290 RDAPYRRDSRRGFSRDNLCKNCKRPGHYAREC----------PNVAICHNCGLPGHIASE 439
Query: 236 CAT------AKGP---SSGGVGGGGCFRCGEVGHLARDCDGGVAVSGGGGNAGRNTCFNC 286
C T K P +S G C CG+ GH AR+C G C NC
Sbjct: 440 CTTKSLCWNCKEPGHMASSCPNEGICHTCGKAGHRARECSAPPMPPG-----DLRLCNNC 604
Query: 287 GKPGHFAREC 296
K GH A EC
Sbjct: 605 YKQGHIAAEC 634
Score = 57.0 bits (136), Expect = 1e-08
Identities = 29/65 (44%), Positives = 33/65 (50%)
Frame = +2
Query: 87 RGFGGWRGGERRNGGGGGGGGGGCYNCGDTGHLARDCHRSNNNGGGGGGAACYNCGDAGH 146
R GG GG R GGGGG C NC GH++RDC G C+NCG GH
Sbjct: 773 RSGGGGGGGGARGGGGGGYRDVVCRNCQQLGHMSRDCM--------GPLMICHNCGGRGH 928
Query: 147 LARDC 151
LA +C
Sbjct: 929 LAYEC 943
>TC216231 similar to UP|Q41188 (Q41188) Glycine-rich protein 2 (GRP2)
(AT4g38680/F20M13_240), partial (42%)
Length = 891
Score = 108 bits (270), Expect = 3e-24
Identities = 73/189 (38%), Positives = 83/189 (43%), Gaps = 5/189 (2%)
Frame = +3
Query: 64 TKAVDVTGPNGAALQPTRKDSAPRGFGGWRGGERRNGGGGGGGGGGCYNCGDTGHLARDC 123
T DV GP+ + +Q TR G G RG GG GG GGGG
Sbjct: 3 TXXXDVXGPDESPVQGTRG-----GGGXXRGSYGXGGGRGGYGGGG-------------- 125
Query: 124 HRSNNNGGGGGGAACYNCGDAGHLARDCNRSNNNSGGGGAGCYNCGDTGHLARDCNRSNN 183
GG GGG Y G G GGGG G
Sbjct: 126 --RGGRGGYGGGGGGYGGGGYG-------------GGGGYG------------------- 203
Query: 184 SGGGGGGGGACYTCGGFGHLARDCMRGGNG----GGGGPGSASCFRCGGIGHMARDC-AT 238
GGGGGGGG CY CG GH+ARDC +GG G GGGG G SC+ CG GH ARDC ++
Sbjct: 204 -GGGGGGGGGCYKCGETGHIARDCSQGGGGGGRYGGGGGGGGSCYNCGESGHFARDCPSS 380
Query: 239 AKGPSSGGV 247
A+ +S GV
Sbjct: 381 AR*YNSTGV 407
Score = 95.9 bits (237), Expect = 2e-20
Identities = 51/114 (44%), Positives = 59/114 (51%), Gaps = 2/114 (1%)
Frame = +3
Query: 185 GGGGGGGGACYTCGGFGHLARDCMRGGNGGGGGPGSASCFRCGGIGHMARDCATAKGPSS 244
G G GGG GG+G R G GGGGG G GG G
Sbjct: 78 GSYGXGGGR----GGYGGGGRGGRGGYGGGGGGYGGGGYGGGGGYG------------GG 209
Query: 245 GGVGGGGCFRCGEVGHLARDCDGGVAVSG--GGGNAGRNTCFNCGKPGHFAREC 296
GG GGGGC++CGE GH+ARDC G G GGG G +C+NCG+ GHFAR+C
Sbjct: 210 GGGGGGGCYKCGETGHIARDCSQGGGGGGRYGGGGGGGGSCYNCGESGHFARDC 371
Score = 94.7 bits (234), Expect = 4e-20
Identities = 51/110 (46%), Positives = 57/110 (51%), Gaps = 4/110 (3%)
Frame = +3
Query: 160 GGGAGCYNCGDTGHLARDCNRSNNSGGGGGGGGACYTCGGFGHLARDCMRGGNGGGGGPG 219
GGG G Y G G R GGGGG GG GG+G GG GGGGG G
Sbjct: 93 GGGRGGYGGGGRG------GRGGYGGGGGGYGG-----GGYGG------GGGYGGGGGGG 221
Query: 220 SASCFRCGGIGHMARDCATAKGP----SSGGVGGGGCFRCGEVGHLARDC 265
C++CG GH+ARDC+ G GG GGG C+ CGE GH ARDC
Sbjct: 222 GGGCYKCGETGHIARDCSQGGGGGGRYGGGGGGGGSCYNCGESGHFARDC 371
Score = 55.8 bits (133), Expect = 2e-08
Identities = 40/116 (34%), Positives = 44/116 (37%)
Frame = +3
Query: 186 GGGGGGGACYTCGGFGHLARDCMRGGNGGGGGPGSASCFRCGGIGHMARDCATAKGPSSG 245
GGGG Y GG RGG GGGG G GG G G
Sbjct: 57 GGGGXXRGSYGXGGG--------RGGYGGGGRGGR------GGYGG-----------GGG 161
Query: 246 GVGGGGCFRCGEVGHLARDCDGGVAVSGGGGNAGRNTCFNCGKPGHFARECIEASG 301
G GGGG GG GGGG G C+ CG+ GH AR+C + G
Sbjct: 162 GYGGGGY--------------GGGGGYGGGGGGGGGGCYKCGETGHIARDCSQGGG 287
>TC227588 similar to PIR|T00837|T00837 glycine-rich protein T13L16.11 -
Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(10%)
Length = 1300
Score = 99.8 bits (247), Expect = 1e-21
Identities = 52/160 (32%), Positives = 72/160 (44%)
Frame = +1
Query: 110 CYNCGDTGHLARDCHRSNNNGGGGGGAACYNCGDAGHLARDCNRSNNNSGGGGAGCYNCG 169
CY CG GH AR C + + C+ C GH A+DC + ++ A C CG
Sbjct: 127 CYVCGCLGHNARQCSKVQD---------CFICKKGGHRAKDCPEKHTSTSKSIAICLKCG 279
Query: 170 DTGHLARDCNRSNNSGGGGGGGGACYTCGGFGHLARDCMRGGNGGGGGPGSASCFRCGGI 229
++GH C N+ CY C GHL C+ N PG SC++CG +
Sbjct: 280 NSGHDIFSCR--NDYSQDDLKEIQCYVCKRLGHLC--CV---NTDDATPGEISCYKCGQL 438
Query: 230 GHMARDCATAKGPSSGGVGGGGCFRCGEVGHLARDCDGGV 269
GH C+ + + G CF+CGE GH AR+C +
Sbjct: 439 GHTGLACSRLRDEITSGATPSSCFKCGEEGHFARECTSSI 558
Score = 94.0 bits (232), Expect = 8e-20
Identities = 57/189 (30%), Positives = 79/189 (41%)
Frame = +1
Query: 108 GGCYNCGDTGHLARDCHRSNNNGGGGGGAACYNCGDAGHLARDCNRSNNNSGGGGAGCYN 167
G C+NCG+ GH A +C CY CG GH AR C++ + C+
Sbjct: 55 GACFNCGEEGHAAVNCSAVKRK------KPCYVCGCLGHNARQCSKVQD--------CFI 192
Query: 168 CGDTGHLARDCNRSNNSGGGGGGGGACYTCGGFGHLARDCMRGGNGGGGGPGSASCFRCG 227
C GH A+DC + S C CG GH C + C+ C
Sbjct: 193 CKKGGHRAKDCPEKHTSTSKSIA--ICLKCGNSGHDIFSCRN--DYSQDDLKEIQCYVCK 360
Query: 228 GIGHMARDCATAKGPSSGGVGGGGCFRCGEVGHLARDCDGGVAVSGGGGNAGRNTCFNCG 287
+GH+ C + G + C++CG++GH C G A ++CF CG
Sbjct: 361 RLGHLC--CVNTDDATPGEIS---CYKCGQLGHTGLACSRLRDEITSG--ATPSSCFKCG 519
Query: 288 KPGHFAREC 296
+ GHFAREC
Sbjct: 520 EEGHFAREC 546
Score = 58.9 bits (141), Expect = 3e-09
Identities = 30/78 (38%), Positives = 38/78 (48%), Gaps = 2/78 (2%)
Frame = +1
Query: 110 CYNCGDTGHLARDCHRSNNNGGGGGGAACYNCGDAGHLARDCNRSNNN--SGGGGAGCYN 167
CY C GHL C N + G +CY CG GH C+R + SG + C+
Sbjct: 346 CYVCKRLGHLC--C--VNTDDATPGEISCYKCGQLGHTGLACSRLRDEITSGATPSSCFK 513
Query: 168 CGDTGHLARDCNRSNNSG 185
CG+ GH AR+C S SG
Sbjct: 514 CGEEGHFARECTSSIKSG 567
Score = 46.2 bits (108), Expect = 2e-05
Identities = 26/77 (33%), Positives = 34/77 (43%)
Frame = +1
Query: 222 SCFRCGGIGHMARDCATAKGPSSGGVGGGGCFRCGEVGHLARDCDGGVAVSGGGGNAGRN 281
+CF CG GH A +C+ K C+ CG +GH AR C +
Sbjct: 58 ACFNCGEEGHAAVNCSAVKRKKP-------CYVCGCLGHNARQC------------SKVQ 180
Query: 282 TCFNCGKPGHFARECIE 298
CF C K GH A++C E
Sbjct: 181 DCFICKKGGHRAKDCPE 231
Score = 31.2 bits (69), Expect = 0.60
Identities = 11/27 (40%), Positives = 15/27 (54%)
Frame = +1
Query: 104 GGGGGGCYNCGDTGHLARDCHRSNNNG 130
G C+ CG+ GH AR+C S +G
Sbjct: 487 GATPSSCFKCGEEGHFARECTSSIKSG 567
>TC227589
Length = 547
Score = 94.4 bits (233), Expect = 6e-20
Identities = 53/171 (30%), Positives = 73/171 (41%), Gaps = 13/171 (7%)
Frame = +2
Query: 108 GGCYNCGDTGHLARDCHRSNNN------GGGGGGAA-------CYNCGDAGHLARDCNRS 154
G C+NCG+ GH A +C + GG G A C+ C GH A+DC
Sbjct: 41 GACFNCGEDGHAAVNCSAAKRKKPCYVCGGLGHNARQCTKAQDCFICKKGGHRAKDCLEK 220
Query: 155 NNNSGGGGAGCYNCGDTGHLARDCNRSNNSGGGGGGGGACYTCGGFGHLARDCMRGGNGG 214
+ + A C CG++GH C N+ CY C GHL C+ N
Sbjct: 221 HTSRSKSVAICLKCGNSGHDMFSCR--NDYSPDDLKEIQCYVCKRVGHLC--CV---NTD 379
Query: 215 GGGPGSASCFRCGGIGHMARDCATAKGPSSGGVGGGGCFRCGEVGHLARDC 265
PG SC++CG +GH C+ + C +CGE GH A++C
Sbjct: 380 DATPGEISCYKCGQLGHTGLACSKLPDEITSAATPSSCCKCGEAGHFAQEC 532
Score = 50.8 bits (120), Expect = 7e-07
Identities = 27/77 (35%), Positives = 35/77 (45%)
Frame = +2
Query: 222 SCFRCGGIGHMARDCATAKGPSSGGVGGGGCFRCGEVGHLARDCDGGVAVSGGGGNAGRN 281
+CF CG GH A +C+ AK C+ CG +GH AR C
Sbjct: 44 ACFNCGEDGHAAVNCSAAKRKKP-------CYVCGGLGHNARQCTKA------------Q 166
Query: 282 TCFNCGKPGHFARECIE 298
CF C K GH A++C+E
Sbjct: 167 DCFICKKGGHRAKDCLE 217
Score = 40.8 bits (94), Expect = 8e-04
Identities = 19/50 (38%), Positives = 24/50 (48%)
Frame = +2
Query: 250 GGCFRCGEVGHLARDCDGGVAVSGGGGNAGRNTCFNCGKPGHFARECIEA 299
G CF CGE GH A +C + C+ CG GH AR+C +A
Sbjct: 41 GACFNCGEDGHAAVNCSAAKR---------KKPCYVCGGLGHNARQCTKA 163
>TC206180 similar to UP|Q8LF59 (Q8LF59) DNA-binding protein, partial (42%)
Length = 655
Score = 90.9 bits (224), Expect = 6e-19
Identities = 58/171 (33%), Positives = 70/171 (40%), Gaps = 13/171 (7%)
Frame = +3
Query: 139 YNCGDAGHLARDCNRSNNNSGGGGAGCYNCGDTGHLARDCNRSNNSGGGGGGGGACYTCG 198
+ CG AGH AR+C+ G C NC GH+A +C AC C
Sbjct: 3 HTCGKAGHRARECSAPPMPPGDLRL-CNNCYKQGHIAAECTNEK----------ACNNCR 149
Query: 199 GFGHLARDCMRGGNGGGGGPGSASCFRCGGIGHMARDCATAK------GPSSGGVGGGG- 251
GHLARDC P C C GH+AR C A G G GGGG
Sbjct: 150 KTGHLARDC----------PNDPICNLCNVSGHVARQCPKANVLGDXXGGGGGARGGGGG 299
Query: 252 ------CFRCGEVGHLARDCDGGVAVSGGGGNAGRNTCFNCGKPGHFAREC 296
C C ++GH++RDC G + + C NCG GH A EC
Sbjct: 300 GYRDVVCRNCQQLGHMSRDCMGPLMI-----------CHNCGGRGHLAYEC 419
Score = 88.2 bits (217), Expect = 4e-18
Identities = 60/171 (35%), Positives = 69/171 (40%), Gaps = 13/171 (7%)
Frame = +3
Query: 111 YNCGDTGHLARDCHRSNNNGGGGGGAACYNCGDAGHLARDCNRSNNNSGGGGAGCYNCGD 170
+ CG GH AR+C S G C NC GH+A +C C NC
Sbjct: 3 HTCGKAGHRAREC--SAPPMPPGDLRLCNNCYKQGHIAAECTNEK--------ACNNCRK 152
Query: 171 TGHLARDCNRSNNSGGGGGGGGACYTCGGFGHLARDCMR-----------GG--NGGGGG 217
TGHLARDC C C GH+AR C + GG GGGGG
Sbjct: 153 TGHLARDCPNDP----------ICNLCNVSGHVARQCPKANVLGDXXGGGGGARGGGGGG 302
Query: 218 PGSASCFRCGGIGHMARDCATAKGPSSGGVGGGGCFRCGEVGHLARDCDGG 268
C C +GHM+RDC GP C CG GHLA +C G
Sbjct: 303 YRDVVCRNCQQLGHMSRDCM---GPLMI------CHNCGGRGHLAYECPSG 428
Score = 53.9 bits (128), Expect = 9e-08
Identities = 27/64 (42%), Positives = 32/64 (49%), Gaps = 7/64 (10%)
Frame = +3
Query: 95 GERRNGGGGGGGGGG-------CYNCGDTGHLARDCHRSNNNGGGGGGAACYNCGDAGHL 147
G+ GGGG GGGG C NC GH++RDC G C+NCG GHL
Sbjct: 252 GDXXGGGGGARGGGGGGYRDVVCRNCQQLGHMSRDCM--------GPLMICHNCGGRGHL 407
Query: 148 ARDC 151
A +C
Sbjct: 408 AYEC 419
Score = 50.1 bits (118), Expect = 1e-06
Identities = 34/107 (31%), Positives = 43/107 (39%), Gaps = 1/107 (0%)
Frame = +3
Query: 195 YTCGGFGHLARDCMRGGNGGGGGPGSAS-CFRCGGIGHMARDCATAKGPSSGGVGGGGCF 253
+TCG GH AR+C + PG C C GH+A +C K C
Sbjct: 3 HTCGKAGHRAREC----SAPPMPPGDLRLCNNCYKQGHIAAECTNEKA----------CN 140
Query: 254 RCGEVGHLARDCDGGVAVSGGGGNAGRNTCFNCGKPGHFARECIEAS 300
C + GHLARDC C C GH AR+C +A+
Sbjct: 141 NCRKTGHLARDCPNDPI------------CNLCNVSGHVARQCPKAN 245
>BU765146 similar to GP|21322711|e pherophorin-dz1 protein {Volvox carteri f.
nagariensis}, partial (25%)
Length = 407
Score = 84.0 bits (206), Expect = 8e-17
Identities = 70/189 (37%), Positives = 70/189 (37%), Gaps = 2/189 (1%)
Frame = +2
Query: 94 GGER--RNGGGGGGGGGGCYNCGDTGHLARDCHRSNNNGGGGGGAACYNCGDAGHLARDC 151
GGER R GGGGGGGGGG G G GGGGGG
Sbjct: 2 GGERFARGGGGGGGGGGGGGGGGGGG------------GGGGGGG--------------- 100
Query: 152 NRSNNNSGGGGAGCYNCGDTGHLARDCNRSNNSGGGGGGGGACYTCGGFGHLARDCMRGG 211
R GGGG G G G GGGGGGGG GGF R RGG
Sbjct: 101 -RGGGGGGGGGGGGGGGGGKG---------GGGGGGGGGGGGGGGGGGFFRGERXXGRGG 250
Query: 212 NGGGGGPGSASCFRCGGIGHMARDCATAKGPSSGGVGGGGCFRCGEVGHLARDCDGGVAV 271
GGGGG G GG G G GG GGGG GG
Sbjct: 251 RGGGGGGGGGGGGGGGGGG---------GGGGEGGGGGGG---------------GGGGG 358
Query: 272 SGGGGNAGR 280
GGGG GR
Sbjct: 359 GGGGGRGGR 385
Score = 80.9 bits (198), Expect = 7e-16
Identities = 62/165 (37%), Positives = 64/165 (38%)
Frame = +1
Query: 86 PRGFGGWRGGERRNGGGGGGGGGGCYNCGDTGHLARDCHRSNNNGGGGGGAACYNCGDAG 145
P G GG GG GGGGGGGGGG R GGGGGG G G
Sbjct: 19 PGGGGGGGGGGGGGGGGGGGGGGG---------------RGGEGGGGGGGGGGGGGGGGG 153
Query: 146 HLARDCNRSNNNSGGGGAGCYNCGDTGHLARDCNRSNNSGGGGGGGGACYTCGGFGHLAR 205
GGGG G G G R+ GGGGGGGG GG G
Sbjct: 154 EGGGGGGGGGGGGGGGGGGGVF*G--GERXREGGERGGGGGGGGGGGG---GGGXGGGGG 318
Query: 206 DCMRGGNGGGGGPGSASCFRCGGIGHMARDCATAKGPSSGGVGGG 250
RGG GGGGG G GG G +G GG GGG
Sbjct: 319 GGGRGGGGGGGGGG-------GGGG---------EGREEGGGGGG 405
Score = 78.2 bits (191), Expect = 4e-15
Identities = 63/166 (37%), Positives = 64/166 (37%), Gaps = 1/166 (0%)
Frame = +3
Query: 87 RGFGGWRGGERRNGGGGGGGGGGCYNCGDTGHLARDCHRSNNNGGGGGGAACYNCGDAGH 146
RG G GG GGGGGGGGGG G G GGGGGG G G
Sbjct: 9 RGLPGGGGGGGGGGGGGGGGGGG----GGGG-------EGGGGGGGGGGGGGGGGGGGG- 152
Query: 147 LARDCNRSNNNSGGGGAGCYNCGDTGHL-ARDCNRSNNSGGGGGGGGACYTCGGFGHLAR 205
R GGGG G G G L R+ GGGGGGGG GG G
Sbjct: 153 -----GRGGGGGGGGGGGGGGGGGGGFLGGREXXGGGGEGGGGGGGGGGGGGGGXGGGGG 317
Query: 206 DCMRGGNGGGGGPGSASCFRCGGIGHMARDCATAKGPSSGGVGGGG 251
GG GGGGG G GG G G GG GGG
Sbjct: 318 GRGEGGGGGGGGGG-------GGGG----------GGEGGGRRGGG 404
Score = 76.6 bits (187), Expect = 1e-14
Identities = 64/174 (36%), Positives = 65/174 (36%)
Frame = +3
Query: 86 PRGFGGWRGGERRNGGGGGGGGGGCYNCGDTGHLARDCHRSNNNGGGGGGAACYNCGDAG 145
P G GG GG GGGGGGGGGG GGGGGG G G
Sbjct: 18 PGGGGGGGGGGGGGGGGGGGGGGG---------------EGGGGGGGGGGGGGGGGGGGG 152
Query: 146 HLARDCNRSNNNSGGGGAGCYNCGDTGHLARDCNRSNNSGGGGGGGGACYTCGGFGHLAR 205
R GGGG G G G L R GGG GGGG GG
Sbjct: 153 ------GRGGGGGGGGGGGGGGGGGGGFLG---GREXXGGGGEGGGGG----GG------ 275
Query: 206 DCMRGGNGGGGGPGSASCFRCGGIGHMARDCATAKGPSSGGVGGGGCFRCGEVG 259
GG GGGGG G GG G +G GG GGGG GE G
Sbjct: 276 ----GGGGGGGGXGGGG----GGRG---------EGGGGGGGGGGGGGGGGEGG 386
Score = 47.0 bits (110), Expect = 1e-05
Identities = 40/100 (40%), Positives = 40/100 (40%)
Frame = +1
Query: 185 GGGGGGGGACYTCGGFGHLARDCMRGGNGGGGGPGSASCFRCGGIGHMARDCATAKGPSS 244
GGGGGGGG GG G GG GGGGG G GG G G
Sbjct: 22 GGGGGGGGG----GGGG--------GGGGGGGGGGR------GGEG----------GGGG 117
Query: 245 GGVGGGGCFRCGEVGHLARDCDGGVAVSGGGGNAGRNTCF 284
GG GGGG GE G GG GGGG G F
Sbjct: 118 GGGGGGGGGGGGEGG------GGGGGGGGGGGGGGGGGVF 219
Score = 42.0 bits (97), Expect = 3e-04
Identities = 29/83 (34%), Positives = 32/83 (37%)
Frame = +1
Query: 82 KDSAPRGFGGWRGGERRNGGGGGGGGGGCYNCGDTGHLARDCHRSNNNGGGGGGAACYNC 141
++ RG GG GG GGG GGGGGG GGGGGG
Sbjct: 238 REGGERGGGGGGGGGGGGGGGXGGGGGG----------------GGRGGGGGGGGG---- 357
Query: 142 GDAGHLARDCNRSNNNSGGGGAG 164
G G R+ GGGG G
Sbjct: 358 GGGGGEGRE-------EGGGGGG 405
Score = 38.5 bits (88), Expect = 0.004
Identities = 32/86 (37%), Positives = 32/86 (37%)
Frame = +2
Query: 209 RGGNGGGGGPGSASCFRCGGIGHMARDCATAKGPSSGGVGGGGCFRCGEVGHLARDCDGG 268
RGG GGGGG G GG G GG GGGG R G G GG
Sbjct: 20 RGGGGGGGGGGG------GG------------GGGGGGGGGGGGGRGGGGGGGGGGGGGG 145
Query: 269 VAVSGGGGNAGRNTCFNCGKPGHFAR 294
GGGG G G G F R
Sbjct: 146 GGGKGGGGGGGGGGGGGGGGGGGFFR 223
>TC210565 similar to GB|AAO27142.1|27904309|AE014017 cold shock protein CspA
{Buchnera aphidicola str. Bp (Baizongia pistaciae);} ,
partial (67%)
Length = 475
Score = 84.0 bits (206), Expect = 8e-17
Identities = 45/97 (46%), Positives = 59/97 (60%), Gaps = 1/97 (1%)
Frame = +3
Query: 4 ERFSGVVQWFNNVKGFGFIKPDDGGEDLFVHQSSIRSD-GYRTLLEGDLVEFSIATGDND 62
+R+ G V+WFN KGFGFI P+DGG DLFVH S+I++D G+RTL EG VEF + D+
Sbjct: 63 QRYKGTVKWFNEEKGFGFITPEDGGSDLFVHYSAIQTDGGFRTLSEGQSVEFLVTQDDSG 242
Query: 63 KTKAVDVTGPNGAALQPTRKDSAPRGFGGWRGGERRN 99
+ AV+VT T S+ G G GG+ N
Sbjct: 243 RAAAVNVT--------TTTVKSSDSGNGENSGGDAAN 329
>TC214106 weakly similar to UP|Q8L685 (Q8L685) Pherophorin-dz1 protein
precursor, partial (22%)
Length = 1296
Score = 80.9 bits (198), Expect = 7e-16
Identities = 69/188 (36%), Positives = 78/188 (40%), Gaps = 11/188 (5%)
Frame = -2
Query: 100 GGGGGGGGGGCYNCGDTGHLARDCHRSNNNGGGGGGAACYNCGDAGHLARDCNRSNNNSG 159
GGGG GGGG Y G G + +GGG GA C GD G +C+ G
Sbjct: 572 GGGGDGGGGELY*TGGGG--------GDGDGGGW*GAGC---GDGG--GGECSSHGGGDG 432
Query: 160 GGGAGCYNCGDTGHLARDCNRSNNSGGGGG----GGGACYTCGGFGHLARDCMRGGNGGG 215
GGG G G G + GGGGG GGG GG G GG GGG
Sbjct: 431 GGGGGSIQGGGGG--GGSIQGGGDGGGGGGE*TGGGGE*TGGGGGGDK*G*TGGGGEGGG 258
Query: 216 GGP-----GSASCFRCGGIGHMARDCATAKGPSSGGVGGGGCFRCGE--VGHLARDCDGG 268
GG G C CGG G + C G ++GG GGGG GE G L + G
Sbjct: 257 GGGECGGGGGGEC--CGGGGEL**YCTGGGGENTGGGGGGGDGGGGEGGGGDLTQYVGEG 84
Query: 269 VAVSGGGG 276
GGGG
Sbjct: 83 GGGEGGGG 60
Score = 77.0 bits (188), Expect = 1e-14
Identities = 68/197 (34%), Positives = 73/197 (36%), Gaps = 5/197 (2%)
Frame = -2
Query: 88 GFGGWRGGERRNGGGGGGGGGGCYNCGDTGHLARDCHRSNNNGGGGGGAACYNCGDAGHL 147
G G GGE + GGG GGGGG + GGGGGG + GD G
Sbjct: 485 GCGDGGGGECSSHGGGDGGGGG----------------GSIQGGGGGGGSIQGGGDGGGG 354
Query: 148 ARDCNRSNNNSGGGGAGCYNCGDTGHLARDCNRSNNSGG--GGGGGGACYTCGGFGHLAR 205
+ GGG G G TG GG GGGGGG C CGG G L
Sbjct: 353 GGE*TGGGGE*TGGGGGGDK*G*TG----GGGEGGGGGGECGGGGGGEC--CGGGGEL** 192
Query: 206 DCMRGG---NGGGGGPGSASCFRCGGIGHMARDCATAKGPSSGGVGGGGCFRCGEVGHLA 262
C GG GGGGG G GG D G GG GGGG
Sbjct: 191 YCTGGGGENTGGGGGGGDGGGGEGGG-----GDLTQYVGEGGGGEGGGG----------G 57
Query: 263 RDCDGGVAVSGGGGNAG 279
+ GG GGG G
Sbjct: 56 GE*TGGGGEGGGGDGGG 6
Score = 68.2 bits (165), Expect = 4e-12
Identities = 62/177 (35%), Positives = 68/177 (38%), Gaps = 13/177 (7%)
Frame = -2
Query: 88 GFGGWRGGERRNGGGGGG--------GGGGCYNCGDTGHLARDCHRSNNNGGGGGGAA-- 137
G GG GG + GGGGGG GGGG G G GGGGGG
Sbjct: 440 GDGGGGGGSIQGGGGGGGSIQGGGDGGGGGGE*TGGGGE*T---------GGGGGGDK*G 288
Query: 138 -CYNCGDAGHLARDCNRSNNNSGGGGAGCYNCGDTGHLARDCNRSN--NSGGGGGGGGAC 194
G+ G +C GGGG C CG G L C N+GGGGGGG
Sbjct: 287 *TGGGGEGGGGGGECG------GGGGGEC--CGGGGEL**YCTGGGGENTGGGGGGGDGG 132
Query: 195 YTCGGFGHLARDCMRGGNGGGGGPGSASCFRCGGIGHMARDCATAKGPSSGGVGGGG 251
GG G L + GG G GGG G + G GG GGGG
Sbjct: 131 GGEGGGGDLTQYVGEGGGGEGGGGGG--------------E*TGGGGEGGGGDGGGG 3
Score = 50.8 bits (120), Expect = 7e-07
Identities = 49/143 (34%), Positives = 54/143 (37%), Gaps = 13/143 (9%)
Frame = -2
Query: 147 LARDCNRSNNNSGGGGAGCYNCGDTGHLARDCN--RSNNSGGGG--------GGGGACYT 196
L ++ N GGGG Y TG N GGGG GGGG
Sbjct: 728 LEEKSSQKNQ*KGGGGGDAYETPMTGGKGPSYTGVGGGNDGGGGEL***TGVGGGGD--- 558
Query: 197 CGGFGHLARDCMRGGNGGGGGPGSASCFRCGG---IGHMARDCATAKGPSSGGVGGGGCF 253
GG G L GG+G GGG A C GG H D G GG GGGG
Sbjct: 557 -GGGGELY*TGGGGGDGDGGGW*GAGCGDGGGGECSSHGGGDGGGGGGSIQGGGGGGGSI 381
Query: 254 RCGEVGHLARDCDGGVAVSGGGG 276
+ G G GG +GGGG
Sbjct: 380 QGGGDGG-----GGGGE*TGGGG 327
>TC228935 similar to UP|Q9FHC2 (Q9FHC2) Arabidopsis thaliana genomic DNA,
chromosome 5, TAC clone:K24M7, partial (36%)
Length = 1224
Score = 74.3 bits (181), Expect = 6e-14
Identities = 38/105 (36%), Positives = 48/105 (45%)
Frame = +1
Query: 110 CYNCGDTGHLARDCHRSNNNGGGGGGAACYNCGDAGHLARDCNRSNNNSGGGGAGCYNCG 169
C C GH A++C + G CYNCG+ GH C G A C+ C
Sbjct: 346 CLRCRRRGHRAKNCPEVLD--GAKDAKYCYNCGENGHALTQCLHPLQEGGTKFAECFVCN 519
Query: 170 DTGHLARDCNRSNNSGGGGGGGGACYTCGGFGHLARDCMRGGNGG 214
GHL+++C N+ G GG C CGG HLA+DC G G
Sbjct: 520 QRGHLSKNC--PQNTHGIYPKGGCCKICGGVTHLAKDCPDKGKSG 648
Score = 67.8 bits (164), Expect = 6e-12
Identities = 39/130 (30%), Positives = 55/130 (42%)
Frame = +1
Query: 107 GGGCYNCGDTGHLARDCHRSNNNGGGGGGAACYNCGDAGHLARDCNRSNNNSGGGGAGCY 166
G C+ C H+A+ C C C GH A++C + + CY
Sbjct: 262 GESCFICKAMDHIAKLCPEKAE---WEKNKICLRCRRRGHRAKNCPEVLDGAKDAKY-CY 429
Query: 167 NCGDTGHLARDCNRSNNSGGGGGGGGACYTCGGFGHLARDCMRGGNGGGGGPGSASCFRC 226
NCG+ GH C GG C+ C GHL+++C + N G P C C
Sbjct: 430 NCGENGHALTQCLHPLQEGGTKFA--ECFVCNQRGHLSKNCPQ--NTHGIYPKGGCCKIC 597
Query: 227 GGIGHMARDC 236
GG+ H+A+DC
Sbjct: 598 GGVTHLAKDC 627
Score = 62.8 bits (151), Expect = 2e-10
Identities = 41/133 (30%), Positives = 57/133 (42%), Gaps = 2/133 (1%)
Frame = +1
Query: 135 GAACYNCGDAGHLARDCNRSNNNSGGGGAGCYNCGDTGHLARDCNRSNNSGGGGGGGGAC 194
G +C+ C H+A+ C C C GH A++C + G C
Sbjct: 262 GESCFICKAMDHIAKLC--PEKAEWEKNKICLRCRRRGHRAKNCPEVLD---GAKDAKYC 426
Query: 195 YTCGGFGHLARDCMRGGNGGGGGPGSASCFRCGGIGHMARDCATAKGPSSGGV--GGGGC 252
Y CG GH C+ GG A CF C GH++++C ++ G+ GG C
Sbjct: 427 YNCGENGHALTQCLHPLQEGG--TKFAECFVCNQRGHLSKNCPQ----NTHGIYPKGGCC 588
Query: 253 FRCGEVGHLARDC 265
CG V HLA+DC
Sbjct: 589 KICGGVTHLAKDC 627
Score = 48.1 bits (113), Expect = 5e-06
Identities = 29/111 (26%), Positives = 42/111 (37%), Gaps = 5/111 (4%)
Frame = +1
Query: 191 GGACYTCGGFGHLARDCMRGGNGGGGGPGSASCFRCGGIGHMARDCATAKGPSSGGVGGG 250
G +C+ C H+A+ C + C RC GH A++C G
Sbjct: 262 GESCFICKAMDHIAKLCPEKAEW----EKNKICLRCRRRGHRAKNCPEVL---DGAKDAK 420
Query: 251 GCFRCGEVGHLARDC-----DGGVAVSGGGGNAGRNTCFNCGKPGHFAREC 296
C+ CGE GH C +GG + CF C + GH ++ C
Sbjct: 421 YCYNCGENGHALTQCLHPLQEGGTKFA---------ECFVCNQRGHLSKNC 546
Score = 43.5 bits (101), Expect = 1e-04
Identities = 27/83 (32%), Positives = 38/83 (45%)
Frame = +1
Query: 215 GGGPGSASCFRCGGIGHMARDCATAKGPSSGGVGGGGCFRCGEVGHLARDCDGGVAVSGG 274
G PG SCF C + H+A+ C + C RC GH A++C V G
Sbjct: 250 GMKPGE-SCFICKAMDHIAKLCPEKAEWEKNKI----CLRCRRRGHRAKNCP---EVLDG 405
Query: 275 GGNAGRNTCFNCGKPGHFARECI 297
+A C+NCG+ GH +C+
Sbjct: 406 AKDA--KYCYNCGENGHALTQCL 468
Score = 28.1 bits (61), Expect = 5.1
Identities = 11/24 (45%), Positives = 13/24 (53%)
Frame = +1
Query: 107 GGGCYNCGDTGHLARDCHRSNNNG 130
GG C CG HLA+DC +G
Sbjct: 577 GGCCKICGGVTHLAKDCPDKGKSG 648
>TC219649 weakly similar to UP|Q95JD1 (Q95JD1) Basic proline-rich protein,
partial (37%)
Length = 750
Score = 73.6 bits (179), Expect = 1e-13
Identities = 71/210 (33%), Positives = 77/210 (35%), Gaps = 1/210 (0%)
Frame = -1
Query: 71 GPNGAALQPTRKDSAPRGFGGWRGGERRNGGGGGGGG-GGCYNCGDTGHLARDCHRSNNN 129
G G + P G+GG GG +GGG GGG GG G G S
Sbjct: 744 GGPGGGVGPGGGGPVGGGYGGGPGGGAGSGGGSYGGGPGGDAGPGGGGPGGSAGPGSGGY 565
Query: 130 GGGGGGAACYNCGDAGHLARDCNRSNNNSGGGGAGCYNCGDTGHLARDCNRSNNSGGGGG 189
GGG GG G+ G S G GG G GD G GGG
Sbjct: 564 GGGPGGDTGPGGGEPG--------SGAGPGSGGYGGGPGGDAGPGGGGPGGGAGPGGGEP 409
Query: 190 GGGACYTCGGFGHLARDCMRGGNGGGGGPGSASCFRCGGIGHMARDCATAKGPSSGGVGG 249
G GA GG+G GG GGG GPG GG G + GPS GG GG
Sbjct: 408 GSGAGPGSGGYG--------GGPGGGVGPGGGG--PGGGAGRGGGGPGSGAGPSGGGPGG 259
Query: 250 GGCFRCGEVGHLARDCDGGVAVSGGGGNAG 279
G GE G GG G GG G
Sbjct: 258 GS--YGGEPG------GGGPGGDGYGGGPG 193
Score = 68.9 bits (167), Expect = 3e-12
Identities = 62/191 (32%), Positives = 69/191 (35%)
Frame = -1
Query: 89 FGGWRGGERRNGGGGGGGGGGCYNCGDTGHLARDCHRSNNNGGGGGGAACYNCGDAGHLA 148
+GG GG GGGG GGG + GG G G Y G G A
Sbjct: 750 YGGGPGGGVGPGGGGPVGGG---------------YGGGPGGGAGSGGGSYGGGPGGD-A 619
Query: 149 RDCNRSNNNSGGGGAGCYNCGDTGHLARDCNRSNNSGGGGGGGGACYTCGGFGHLARDCM 208
S G G+G Y G G GGG G GA GG+G
Sbjct: 618 GPGGGGPGGSAGPGSGGYGGGPGG--------DTGPGGGEPGSGAGPGSGGYG------- 484
Query: 209 RGGNGGGGGPGSASCFRCGGIGHMARDCATAKGPSSGGVGGGGCFRCGEVGHLARDCDGG 268
GG GG GPG GG G + + GP SGG GGG G VG GG
Sbjct: 483 -GGPGGDAGPGGGG--PGGGAGPGGGEPGSGAGPGSGGYGGG---PGGGVGPGGGGPGGG 322
Query: 269 VAVSGGGGNAG 279
GGG +G
Sbjct: 321 AGRGGGGPGSG 289
Score = 66.2 bits (160), Expect = 2e-11
Identities = 62/184 (33%), Positives = 72/184 (38%), Gaps = 4/184 (2%)
Frame = -1
Query: 71 GPNGAALQPTRKDSAPRGFGGWRGGERRNGGGGGGGGGGCYNCGDTGHLARDCHRSNNNG 130
GP G+A G+GG GG+ GGG G G G + G G D + G
Sbjct: 603 GPGGSA------GPGSGGYGGGPGGDTGPGGGEPGSGAGPGSGGYGGGPGGD---AGPGG 451
Query: 131 GGGGGAACYNCGDAGHLARDCNRSNNNSGGGGAGCYNCGDTGHLAR----DCNRSNNSGG 186
GG GG A G+ G A + GGG G G G R + + SGG
Sbjct: 450 GGPGGGAGPGGGEPGSGAGPGSGGYGGGPGGGVGPGGGGPGGGAGRGGGGPGSGAGPSGG 271
Query: 187 GGGGGGACYTCGGFGHLARDCMRGGNGGGGGPGSASCFRCGGIGHMARDCATAKGPSSGG 246
G GGG GG G GG+G GGGPG GG G GP GG
Sbjct: 270 GPGGGSYGGEPGGGG-------PGGDGYGGGPG-------GGAG------PDGGGPKDGG 151
Query: 247 VGGG 250
GGG
Sbjct: 150 FGGG 139
Score = 43.9 bits (102), Expect = 9e-05
Identities = 46/140 (32%), Positives = 50/140 (34%), Gaps = 9/140 (6%)
Frame = -1
Query: 160 GGGAGCYNCGDTGHLARDCNRSNNSGGGGGGGGACYTCGGFGHLARDCMRGGNGGGG--- 216
GGG G G G GGG G GG Y G G G GGGG
Sbjct: 735 GGGVGPGGGGPVGG-----GYGGGPGGGAGSGGGSYGGGPGGD-------AGPGGGGPGG 592
Query: 217 --GPGSASC--FRCGGIGHMARDCATAKGPSSGGVGGGGCFRCGEVGHLARDCDGGVAVS 272
GPGS G G + + GP SGG GGG G+ G GG
Sbjct: 591 SAGPGSGGYGGGPGGDTGPGGGEPGSGAGPGSGGYGGG---PGGDAGPGGGGPGGGAGPG 421
Query: 273 GG--GGNAGRNTCFNCGKPG 290
GG G AG + G PG
Sbjct: 420 GGEPGSGAGPGSGGYGGGPG 361
>TC214476 weakly similar to UP|Q8L685 (Q8L685) Pherophorin-dz1 protein
precursor, partial (14%)
Length = 642
Score = 69.7 bits (169), Expect = 2e-12
Identities = 59/168 (35%), Positives = 67/168 (39%), Gaps = 3/168 (1%)
Frame = -3
Query: 87 RGFGGWRGGERRNGGGGGGGGGGCYNCGDTGHLARDCHRSNNNGGGGGGAACYNCGDAGH 146
+G GG+ GG + GGG GGGG + G +G A S + GGG GG
Sbjct: 601 KGGGGYGGGGSQGGGGSAGGGG---S*GGSGGSAGGGEGSGSQGGGSGGGG--------- 458
Query: 147 LARDCNRSNNNSGGGGAGCYNCGDTGHLARDCNRSNNSGGGGGGGGACYTCGGFGHLARD 206
S G G G Y G +G +SGGGGGGGG GG G
Sbjct: 457 -------SQGGGSGSGGGGYGGGGSG----GSEGGGSSGGGGGGGG-----GGGG----- 341
Query: 207 CMRGGNGGGGGPGSASCFRCGGIGHMARDC---ATAKGPSSGGVGGGG 251
GG GGGG G GGIG D KG GG GGG
Sbjct: 340 ---GGYGGGGDKG-------GGIGGDKGDYHHGKGGKGDKDGGDKGGG 227
Score = 68.6 bits (166), Expect = 3e-12
Identities = 59/177 (33%), Positives = 66/177 (36%), Gaps = 1/177 (0%)
Frame = -3
Query: 100 GGGGGGGGGGCYNCGDTGHLARDCHRSNNNGGGGGGAACYNCGDAGHLARDCNRSNNNSG 159
GGGG GGGG S GG GGG + G AG + G
Sbjct: 634 GGGGSAGGGG----------------SKGGGGYGGGGSQGGGGSAGGGGS*GGSGGSAGG 503
Query: 160 GGGAGCYNCGDTGHLARDCNRSNNSGGGGGGGGACYTCGGFGHLARDCMRGGNGGGGGPG 219
G G+G G G + GGG G GG Y GG G GG GGGGG G
Sbjct: 502 GEGSGSQGGGSGG--------GGSQGGGSGSGGGGYGGGGSGGSEGGGSSGGGGGGGGGG 347
Query: 220 SASCFRCGGIGHMARDCATAKGPSSGGVGGG-GCFRCGEVGHLARDCDGGVAVSGGG 275
GG G G GG+GG G + G+ G D DGG GGG
Sbjct: 346 GG-----GGYG--------GGGDKGGGIGGDKGDYHHGKGG--KGDKDGGDKGGGGG 221
Score = 50.8 bits (120), Expect = 7e-07
Identities = 35/101 (34%), Positives = 41/101 (39%), Gaps = 3/101 (2%)
Frame = -3
Query: 182 NNSGGGGGGGGACYTCGGFGHLARDCMRGGNGGGGGPGSASCFRCGGIGHMARDCATAKG 241
+ GGG GGG GG+G G GGGG G + GG G ++ + G
Sbjct: 640 SEGGGGSAGGGGSKGGGGYGGGGSQGGGGSAGGGGS*GGSGGSAGGGEGSGSQGGGSGGG 461
Query: 242 PSSG---GVGGGGCFRCGEVGHLARDCDGGVAVSGGGGNAG 279
S G G GGGG G G GG GGGG G
Sbjct: 460 GSQGGGSGSGGGGYGGGGSGGSEGGGSSGGGGGGGGGGGGG 338
Score = 49.7 bits (117), Expect = 2e-06
Identities = 35/106 (33%), Positives = 36/106 (33%)
Frame = -3
Query: 90 GGWRGGERRNGGGGGGGGGGCYNCGDTGHLARDCHRSNNNGGGGGGAACYNCGDAGHLAR 149
GG GG GGG G GGGG G G GGGGGG Y G
Sbjct: 481 GGGSGGGGSQGGGSGSGGGGYGGGGSGGSEGGGSSGGGGGGGGGGGGGGYGGG------- 323
Query: 150 DCNRSNNNSGGGGAGCYNCGDTGHLARDCNRSNNSGGGGGGGGACY 195
GGG G GD G + GG GGG Y
Sbjct: 322 -------GDKGGGIG----GDKGDYHHGKGGKGDKDGGDKGGGGGY 218
Score = 46.6 bits (109), Expect = 1e-05
Identities = 42/134 (31%), Positives = 48/134 (35%)
Frame = -3
Query: 84 SAPRGFGGWRGGERRNGGGGGGGGGGCYNCGDTGHLARDCHRSNNNGGGGGGAACYNCGD 143
SA G G G GGG GGG G G G + ++GGGGGG
Sbjct: 514 SAGGGEGSGSQGGGSGGGGSQGGGSGSGGGGYGGGGSGGSEGGGSSGGGGGG-------- 359
Query: 144 AGHLARDCNRSNNNSGGGGAGCYNCGDTGHLARDCNRSNNSGGGGGGGGACYTCGGFGHL 203
GGGG G Y G + GGG GG Y G G
Sbjct: 358 --------------GGGGGGGGYGGG------------GDKGGGIGGDKGDYHHGKGGKG 257
Query: 204 ARDCMRGGNGGGGG 217
+D GG+ GGGG
Sbjct: 256 DKD---GGDKGGGG 224
Score = 46.2 bits (108), Expect = 2e-05
Identities = 37/119 (31%), Positives = 41/119 (34%)
Frame = -3
Query: 71 GPNGAALQPTRKDSAPRGFGGWRGGERRNGGGGGGGGGGCYNCGDTGHLARDCHRSNNNG 130
G G Q S G+GG G GG GGGGGG G
Sbjct: 475 GSGGGGSQGGGSGSGGGGYGGGGSGGSEGGGSSGGGGGG--------------------G 356
Query: 131 GGGGGAACYNCGDAGHLARDCNRSNNNSGGGGAGCYNCGDTGHLARDCNRSNNSGGGGG 189
GGGGG GD G GG G Y+ G G +D + GGGGG
Sbjct: 355 GGGGGGGYGGGGDKG-----------GGIGGDKGDYHHGKGGKGDKD---GGDKGGGGG 221
>TC210023 weakly similar to UP|Q868B4 (Q868B4) C. elegans GRL-25 protein
(Corresponding sequence ZK643.8), partial (12%)
Length = 819
Score = 63.2 bits (152), Expect = 1e-10
Identities = 67/230 (29%), Positives = 75/230 (32%), Gaps = 7/230 (3%)
Frame = +3
Query: 73 NGAALQPTRKDSAPRGFG-------GWRGGERRNGGGGGGGGGGCYNCGDTGHLARDCHR 125
NG L T + G G G +GG G G GGGC N +G
Sbjct: 18 NGTGLSSTNMCNEDEGITYKSEISIGGCGNAVESGGCGAGCGGGCGNLIKSG-------- 173
Query: 126 SNNNGGGGGGAACYNCGDAGHLARDCNRSNNNSGGGGAGCYNCGDTGHLARDCNRSNNSG 185
G GG GA C G G+L + + G GAGC CG C SG
Sbjct: 174 ----GCGGCGAGCG--GGCGNLIK-----SGGCGRCGAGCGGCGG------GCGNLIKSG 302
Query: 186 GGGGGGGACYTCGGFGHLARDCMRGGNGGGGGPGSASCFRCGGIGHMARDCATAKGPSSG 245
G GG G C GGG G G S + GG G C G
Sbjct: 303 GCGGCGAGC------------------GGGCGGGCGSMLKSGGCGGCGGGC--------G 404
Query: 246 GVGGGGCFRCGEVGHLARDCDGGVAVSGGGGNAGRNTCFNCGKPGHFARE 295
G GGG C + SG GG G CG HF E
Sbjct: 405 GCGGG--------------CGNRLESSGCGGCGGDLMDSKCGIDEHFNEE 512
Score = 27.3 bits (59), Expect = 8.7
Identities = 21/71 (29%), Positives = 26/71 (36%)
Frame = +3
Query: 88 GFGGWRGGERRNGGGGGGGGGGCYNCGDTGHLARDCHRSNNNGGGGGGAACYNCGDAGHL 147
G GG GG + GG GG CG G + S+ GG GG CG H
Sbjct: 324 GCGGGCGGGCGSMLKSGGCGGCGGGCGGCGGGCGNRLESSGCGGCGGDLMDSKCGIDEHF 503
Query: 148 ARDCNRSNNNS 158
+ N +
Sbjct: 504 NEELKYVNEEA 536
>TC206177 similar to UP|Q41188 (Q41188) Glycine-rich protein 2 (GRP2)
(AT4g38680/F20M13_240), partial (22%)
Length = 406
Score = 62.0 bits (149), Expect = 3e-10
Identities = 27/53 (50%), Positives = 32/53 (59%)
Frame = +1
Query: 89 FGGWRGGERRNGGGGGGGGGGCYNCGDTGHLARDCHRSNNNGGGGGGAACYNC 141
+GG GG GGGGGGGGG CY+CG++GH ARDC S GG +C
Sbjct: 43 YGGGGGGRYGGGGGGGGGGGSCYSCGESGHFARDCPSSAR*NYCYGGLCYADC 201
Score = 61.6 bits (148), Expect = 4e-10
Identities = 33/72 (45%), Positives = 38/72 (51%), Gaps = 3/72 (4%)
Frame = +1
Query: 98 RNGGGGG---GGGGGCYNCGDTGHLARDCHRSNNNGGGGGGAACYNCGDAGHLARDCNRS 154
R GGGGG GGGGG Y G GGGGGG +CY+CG++GH ARDC S
Sbjct: 19 RGGGGGGRYGGGGGGRYGGG--------------GGGGGGGGSCYSCGESGHFARDCPSS 156
Query: 155 NNNSGGGGAGCY 166
+ G CY
Sbjct: 157 AR*NYCYGGLCY 192
Score = 60.8 bits (146), Expect = 7e-10
Identities = 33/69 (47%), Positives = 36/69 (51%), Gaps = 2/69 (2%)
Frame = +1
Query: 159 GGGGAGCYNCGDTGHLARDCNRSNNSGGGGGGGGACYTCGGFGHLARDCMRGG--NGGGG 216
GGGG G Y G G R GGGGGGGG+CY+CG GH ARDC N G
Sbjct: 22 GGGGGGRYGGGGGG-------RYGGGGGGGGGGGSCYSCGESGHFARDCPSSAR*NYCYG 180
Query: 217 GPGSASCFR 225
G A CF+
Sbjct: 181 GLCYADCFK 207
Score = 57.0 bits (136), Expect = 1e-08
Identities = 31/69 (44%), Positives = 36/69 (51%), Gaps = 2/69 (2%)
Frame = +1
Query: 188 GGGGGACYTCGGFGHLARDCMRGGNGGGGGPGSASCFRCGGIGHMARDCATAK--GPSSG 245
GGGGG Y GG G R G GGGGG G SC+ CG GH ARDC ++ G
Sbjct: 22 GGGGGGRYGGGGGG-------RYGGGGGGGGGGGSCYSCGESGHFARDCPSSAR*NYCYG 180
Query: 246 GVGGGGCFR 254
G+ CF+
Sbjct: 181 GLCYADCFK 207
Score = 47.8 bits (112), Expect = 6e-06
Identities = 25/55 (45%), Positives = 26/55 (46%)
Frame = +1
Query: 211 GNGGGGGPGSASCFRCGGIGHMARDCATAKGPSSGGVGGGGCFRCGEVGHLARDC 265
G GGGG G R GG G GG GGG C+ CGE GH ARDC
Sbjct: 22 GGGGGGRYGGGGGGRYGGGG-------------GGGGGGGSCYSCGESGHFARDC 147
Score = 47.4 bits (111), Expect = 8e-06
Identities = 23/49 (46%), Positives = 28/49 (56%)
Frame = +1
Query: 248 GGGGCFRCGEVGHLARDCDGGVAVSGGGGNAGRNTCFNCGKPGHFAREC 296
GGGG R G G GG GGGG G +C++CG+ GHFAR+C
Sbjct: 22 GGGGGGRYGGGG-------GGRYGGGGGGGGGGGSCYSCGESGHFARDC 147
>TC214303 similar to UP|Q9T0K5 (Q9T0K5) Extensin-like protein, partial (18%)
Length = 1014
Score = 61.2 bits (147), Expect = 5e-10
Identities = 51/137 (37%), Positives = 55/137 (39%), Gaps = 5/137 (3%)
Frame = -1
Query: 88 GFGGWRGGERRNGGGGGGGGGG----CYNCGDTGHLARDCHRSNNNGGGGGGAACYNCGD 143
G G GGE GGGGG G G CGD G +C GGGGGG + GD
Sbjct: 369 GGGDGGGGELY*IGGGGGDGDGGGW*GAECGDGG--GGECSSQGGGGGGGGGDSIQGGGD 196
Query: 144 AGHLARDCNRSNNNSGGGGAGCYNCGDT-GHLARDCNRSNNSGGGGGGGGACYTCGGFGH 202
G + GGG G G T G +C GG GGGGG Y GG G
Sbjct: 195 GGGGGGE*TGGGGE*TGGGGGGDK*G*TGGGGGGEC----GGGGDGGGGGGEYVGGGAG- 31
Query: 203 LARDCMRGGNGGGGGPG 219
G GGGG G
Sbjct: 30 -------GEYTGGGGGG 1
Score = 60.1 bits (144), Expect = 1e-09
Identities = 64/188 (34%), Positives = 70/188 (37%)
Frame = -1
Query: 95 GERRNGGGGGGGGGGCYNCGDTGHLARDCHRSNNNGGGGGGAACYNCGDAGHLARDCNRS 154
G G GGG GGG TG GG GGG Y G G
Sbjct: 444 GPSYTGAGGGDDGGGGEL***TGV---------GGGGDGGGGELY*IGGGG--------- 319
Query: 155 NNNSGGGGAGCYNCGDTGHLARDCNRSNNSGGGGGGGGACYTCGGFGHLARDCMRGGNGG 214
+ GGG G CGD G +C S+ GGGGGGGG D ++GG G
Sbjct: 318 GDGDGGGW*GA-ECGDGG--GGEC--SSQGGGGGGGGG-------------DSIQGGGDG 193
Query: 215 GGGPGSASCFRCGGIGHMARDCATAKGPSSGGVGGGGCFRCGEVGHLARDCDGGVAVSGG 274
GGG G + GG G G G GGGG CG G D GG G
Sbjct: 192 GGGGGE*T----GGGGE*TG--GGGGGDK*G*TGGGGGGECGGGG----DGGGGGGEYVG 43
Query: 275 GGNAGRNT 282
GG G T
Sbjct: 42 GGAGGEYT 19
Score = 59.7 bits (143), Expect = 2e-09
Identities = 60/180 (33%), Positives = 65/180 (35%), Gaps = 3/180 (1%)
Frame = -1
Query: 100 GGGGGGGGGGCYNCGDTGHLARDCHRSNNNGGGGGGAACYNCGDAGHLARDCNRSNNNSG 159
GGGG GGGG Y G G + +GGG GA CGD G +C+ G
Sbjct: 372 GGGGDGGGGELY*IGGGG--------GDGDGGGW*GA---ECGDGG--GGECSSQGGGGG 232
Query: 160 GGGAGCYNCGDTGHLARDCNRSNNSGGGGGGGGACYTCGGFGHLARDCMRGGNGGG---G 216
GGG GD+ GGG GGGG GG G GG GGG G
Sbjct: 231 GGG------GDS-----------IQGGGDGGGGGGE*TGGGGE*T-----GGGGGGDK*G 118
Query: 217 GPGSASCFRCGGIGHMARDCATAKGPSSGGVGGGGCFRCGEVGHLARDCDGGVAVSGGGG 276
G CGG GG GGGG G GG GGGG
Sbjct: 117 *TGGGGGGECGG----------------GGDGGGGG------GEYVGGGAGGEYTGGGGG 4
Score = 50.1 bits (118), Expect = 1e-06
Identities = 50/167 (29%), Positives = 57/167 (33%), Gaps = 28/167 (16%)
Frame = -1
Query: 138 CYNCGDAGHLARDCNRSNNNSGGGGAGCYNCGDTGHLARDCNRSNN-------------- 183
CY G+ L ++ N GGGG Y TG +
Sbjct: 549 CYGRGEG--LEEKSSQKNQ*KGGGGGDAYETPMTGGKGPSYTGAGGGDDGGGGEL***TG 376
Query: 184 -SGGGGGGGGACYTCG-------GFGHLARDCMRGG------NGGGGGPGSASCFRCGGI 229
GGG GGGG Y G G G +C GG GGGGG G + GG
Sbjct: 375 VGGGGDGGGGELY*IGGGGGDGDGGGW*GAECGDGGGGECSSQGGGGGGGGGDSIQGGGD 196
Query: 230 GHMARDCATAKGPSSGGVGGGGCFRCGEVGHLARDCDGGVAVSGGGG 276
G T G G GGGG G +C GG GGGG
Sbjct: 195 GGGGGGE*TGGGGE*TGGGGGGDK*G*TGGGGGGECGGGGDGGGGGG 55
>TC204079 similar to UP|Q9SWA8 (Q9SWA8) Glycine-rich RNA-binding protein,
complete
Length = 748
Score = 58.9 bits (141), Expect = 3e-09
Identities = 46/122 (37%), Positives = 49/122 (39%)
Frame = +1
Query: 96 ERRNGGGGGGGGGGCYNCGDTGHLARDCHRSNNNGGGGGGAACYNCGDAGHLARDCNRSN 155
+ R GGGGGGGGGG Y G R GGGG G G G+ NRS
Sbjct: 319 QSRGGGGGGGGGGGGYGGG----------RGGGYGGGGRG------GGGGY-----NRSG 435
Query: 156 NNSGGGGAGCYNCGDTGHLARDCNRSNNSGGGGGGGGACYTCGGFGHLARDCMRGGNGGG 215
G GG G Y G GG GGG Y GG R RGG+GG
Sbjct: 436 GGGGYGGGGGYGGG-------------GGGGYGGGRDRGYGGGG----DRGYSRGGDGGD 564
Query: 216 GG 217
GG
Sbjct: 565 GG 570
Score = 48.1 bits (113), Expect = 5e-06
Identities = 37/100 (37%), Positives = 41/100 (41%)
Frame = +1
Query: 180 RSNNSGGGGGGGGACYTCGGFGHLARDCMRGGNGGGGGPGSASCFRCGGIGHMARDCATA 239
+S GGGGGGGG Y GG G GG GGGG G R GG G
Sbjct: 319 QSRGGGGGGGGGGGGYG-GGRG--------GGYGGGGRGGGGGYNRSGGGGGYG------ 453
Query: 240 KGPSSGGVGGGGCFRCGEVGHLARDCDGGVAVSGGGGNAG 279
G G GGGG + G D G + G GG+ G
Sbjct: 454 -GGGGYGGGGGGGYGGGRDRGYGGGGDRGYSRGGDGGDGG 570
Score = 48.1 bits (113), Expect = 5e-06
Identities = 36/103 (34%), Positives = 42/103 (39%)
Frame = +1
Query: 152 NRSNNNSGGGGAGCYNCGDTGHLARDCNRSNNSGGGGGGGGACYTCGGFGHLARDCMRGG 211
+R GGGG G Y G G GG GGGGG + GG G+ GG
Sbjct: 322 SRGGGGGGGGGGGGYGGGRGG--------GYGGGGRGGGGGYNRSGGGGGYGGGGGYGGG 477
Query: 212 NGGGGGPGSASCFRCGGIGHMARDCATAKGPSSGGVGGGGCFR 254
GGG G G + GG +G S GG GG G +R
Sbjct: 478 GGGGYGGGRDRGYGGGG----------DRGYSRGGDGGDGGWR 576
Score = 48.1 bits (113), Expect = 5e-06
Identities = 38/101 (37%), Positives = 45/101 (43%)
Frame = +1
Query: 179 NRSNNSGGGGGGGGACYTCGGFGHLARDCMRGGNGGGGGPGSASCFRCGGIGHMARDCAT 238
N + + GGGGGGGG GG+G GG GGG G G R GG G+
Sbjct: 310 NEAQSRGGGGGGGGG---GGGYG--------GGRGGGYGGGG----RGGGGGY------- 423
Query: 239 AKGPSSGGVGGGGCFRCGEVGHLARDCDGGVAVSGGGGNAG 279
+ GG GGGG + G G D G GGGG+ G
Sbjct: 424 NRSGGGGGYGGGGGYGGGGGGGYGGGRDRGY---GGGGDRG 537
Score = 41.6 bits (96), Expect = 4e-04
Identities = 25/58 (43%), Positives = 28/58 (48%)
Frame = +1
Query: 88 GFGGWRGGERRNGGGGGGGGGGCYNCGDTGHLARDCHRSNNNGGGGGGAACYNCGDAG 145
G G GG R+GGGGG GGGG Y G G + G GGGG Y+ G G
Sbjct: 397 GGRGGGGGYNRSGGGGGYGGGGGYGGGGGGGYG----GGRDRGYGGGGDRGYSRGGDG 558
Score = 36.2 bits (82), Expect = 0.019
Identities = 24/62 (38%), Positives = 25/62 (39%), Gaps = 7/62 (11%)
Frame = +1
Query: 88 GFGGWRGGERRNGGGG-------GGGGGGCYNCGDTGHLARDCHRSNNNGGGGGGAACYN 140
G GG G R GGGG GGGGGG Y G R + GG GG N
Sbjct: 400 GRGGGGGYNRSGGGGGYGGGGGYGGGGGGGYGGGRDRGYGGGGDRGYSRGGDGGDGGWRN 579
Query: 141 CG 142
G
Sbjct: 580 *G 585
Score = 27.7 bits (60), Expect = 6.6
Identities = 22/66 (33%), Positives = 26/66 (39%), Gaps = 7/66 (10%)
Frame = +1
Query: 234 RDCATAKGPSSGGVGGG-------GCFRCGEVGHLARDCDGGVAVSGGGGNAGRNTCFNC 286
R+ + S GG GGG G R G G R GG SGGGG G +
Sbjct: 295 RNITVNEAQSRGGGGGGGGGGGGYGGGRGGGYGGGGRGGGGGYNRSGGGGGYGGGGGYGG 474
Query: 287 GKPGHF 292
G G +
Sbjct: 475 GGGGGY 492
>TC231868 similar to UP|Q93424 (Q93424) C. elegans GRL-23 protein
(Corresponding sequence E02A10.2), partial (23%)
Length = 790
Score = 54.7 bits (130), Expect = 5e-08
Identities = 46/133 (34%), Positives = 50/133 (37%), Gaps = 5/133 (3%)
Frame = +1
Query: 100 GGGGGGGGGGCYNCGDTGHLARDCHRSNNNGGGGG----GAACYNCGDAGHLARDCNRSN 155
GG G G GGGC N +G C GGG G G C CG
Sbjct: 181 GGCGAGCGGGCGNMIKSGGCGSGC------GGGCGNLIKGGGCGGCG------------- 303
Query: 156 NNSGGGGAGCYNCGDTGHLARDCNRSNNSGGGGGGGGACYTC-GGFGHLARDCMRGGNGG 214
GG G GC N +G C GGG GG C C GG G+ GG GG
Sbjct: 304 ---GGCGGGCGNLIKSGGCGAGC--------GGGCGGGCGGCGGGCGNRLESSGCGGCGG 450
Query: 215 GGGPGSASCFRCG 227
G G G +CG
Sbjct: 451 GCG-GDLMDSKCG 486
Score = 47.0 bits (110), Expect = 1e-05
Identities = 37/116 (31%), Positives = 45/116 (37%), Gaps = 6/116 (5%)
Frame = +1
Query: 178 CNRSNNSGGGGGGGGACYTCGGFGHLARDCMRGGNGGGGGPGSASCFRCGGIGHMARDCA 237
C + SGG GG G C GG G++ + GG G G G G + + GG G
Sbjct: 151 CGNAMESGGCGGCGAGCG--GGCGNMIKS---GGCGSGCGGGCGNLIKGGGCG------- 294
Query: 238 TAKGPSSGGVGGGGCFRCGEVGHLARDCDGGV--AVSGGGGNAGR----NTCFNCG 287
GG GGGC + G C GG G GG G + C CG
Sbjct: 295 -----GCGGGCGGGCGNLIKSGGCGAGCGGGCGGGCGGCGGGCGNRLESSGCGGCG 447
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.317 0.144 0.482
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,256,190
Number of Sequences: 63676
Number of extensions: 364518
Number of successful extensions: 17264
Number of sequences better than 10.0: 810
Number of HSP's better than 10.0 without gapping: 4230
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 10005
length of query: 301
length of database: 12,639,632
effective HSP length: 97
effective length of query: 204
effective length of database: 6,463,060
effective search space: 1318464240
effective search space used: 1318464240
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0197.11


