

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0133.10
(382 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
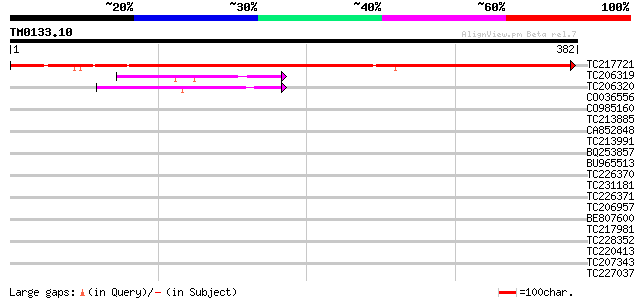
Score E
Sequences producing significant alignments: (bits) Value
TC217721 similar to GB|AAR24767.1|38604048|BT010989 At5g21280 {A... 524 e-149
TC206319 similar to UP|Q9Y0C9 (Q9Y0C9) Ras interacting protein R... 44 1e-04
TC206320 similar to UP|Q9W4F9 (Q9W4F9) CG32772-PA, partial (10%) 43 2e-04
CO036556 40 0.001
CO985160 40 0.001
TC213885 weakly similar to UP|Q9VUB7 (Q9VUB7) CG32133-PA, partia... 40 0.002
CA852848 37 0.011
TC213991 similar to UP|Q6H976 (Q6H976) STYLOSA protein, partial ... 37 0.015
BQ253857 36 0.025
BU965513 35 0.043
TC226370 similar to UP|Q9M0H8 (Q9M0H8) Predicted proline-rich pr... 35 0.043
TC231181 weakly similar to UP|Q86PD9 (Q86PD9) RE65015p, partial ... 35 0.043
TC226371 similar to UP|Q9M0H8 (Q9M0H8) Predicted proline-rich pr... 35 0.056
TC206957 weakly similar to UP|Q9LUI1 (Q9LUI1) Extensin protein-l... 35 0.074
BE807600 similar to GP|24461867|gb CTV.22 {Poncirus trifoliata},... 34 0.096
TC217981 similar to UP|Q9VSK3 (Q9VSK3) CG7015-PA, partial (3%) 34 0.13
TC228352 weakly similar to GB|AAP13389.1|30023712|BT006281 At1g5... 33 0.16
TC220413 similar to UP|Q9LF51 (Q9LF51) Glutamine-rich protein, p... 33 0.28
TC207343 similar to GB|AAN31116.1|23506209|AY149962 At3g06130/F2... 33 0.28
TC227037 homologue to UP|MSI4_ARATH (O22607) WD-40 repeat protei... 33 0.28
>TC217721 similar to GB|AAR24767.1|38604048|BT010989 At5g21280 {Arabidopsis
thaliana;} , partial (15%)
Length = 1636
Score = 524 bits (1350), Expect = e-149
Identities = 279/393 (70%), Positives = 317/393 (79%), Gaps = 12/393 (3%)
Frame = +2
Query: 1 MRVRSSFKQESNRGMELNKLSRNLEEEPPFSGAYMRSLVKQL---RTND------QGCVV 51
MRV ++ +GMEL++LS+ EEPP SGA +R LVKQL RT D Q CVV
Sbjct: 239 MRVDPI*SKKRKKGMELDRLSKL--EEPPLSGACIRGLVKQLTTSRTKDFMNPKYQNCVV 412
Query: 52 NSDASSHGQNLTKHGKIGKACQSQQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARRE 111
N S HGQN TKHGK+ Q+QQS+Q PQQ KKQVRRRLHTSRPYQERLLNMAEARRE
Sbjct: 413 NGGVS-HGQNSTKHGKVADTRQAQQSSQ-PQQQKKQVRRRLHTSRPYQERLLNMAEARRE 586
Query: 112 IVTALKFHRASMKEASEQQQQQQLQQQQQQRAPDSPQLSQNPSFDQDGRYKSRRNPRIYP 171
IVTALKFHRA+MKEASEQ+QQQQ Q+QQQR S Q SQ PSF+QDG++KSRRNPRIYP
Sbjct: 587 IVTALKFHRAAMKEASEQKQQQQQAQEQQQRPSVSLQPSQFPSFNQDGKFKSRRNPRIYP 766
Query: 172 SCNTNFSSYMG-DFSSYPPAFVPNSYPVPVASPIAPPPLMAENPNFILPNQPLGLNLNFH 230
+C T FSSYM D S PP VPNSY P ASPI PPPLMAENPNF+LPNQ LGLNLN
Sbjct: 767 ACTTKFSSYMDMDDLSQPPPLVPNSYTWPAASPITPPPLMAENPNFVLPNQTLGLNLNLQ 946
Query: 231 DFNNLDATIHLSNTSLSSNSFSSATSSS--QEVPSVELSQGDGISSLVNSTESNAASQVN 288
DFNNLDAT+HL+N+S SS+S+SSATSSS QE+PSV +SQG+G SSLV++ ESNAA+QV
Sbjct: 947 DFNNLDATLHLNNSS-SSSSYSSATSSSPPQELPSVGISQGEGFSSLVDTIESNAATQVT 1123
Query: 289 AGLHAAMDEEAIAEIRSLGEQYQMEWNDTMNLVKSACWFKFLRNMEHGAPEANTEGDAYH 348
GLH AMD+E +AE+RSLGEQYQMEWNDTMNLVKSACWFK+L+NMEH A EANT AY+
Sbjct: 1124GGLHTAMDDEGMAEMRSLGEQYQMEWNDTMNLVKSACWFKYLKNMEHRAHEANTVDVAYN 1303
Query: 349 NFDQPLEFPAWLNANESCLDQCSVEYFQNSTLP 381
NFDQ LEFPAWLNANESCL+QCSV+YFQ S+LP
Sbjct: 1304NFDQLLEFPAWLNANESCLEQCSVDYFQESSLP 1402
>TC206319 similar to UP|Q9Y0C9 (Q9Y0C9) Ras interacting protein RIPA, partial
(5%)
Length = 1428
Score = 43.9 bits (102), Expect = 1e-04
Identities = 35/118 (29%), Positives = 52/118 (43%), Gaps = 4/118 (3%)
Frame = +3
Query: 73 QSQQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARR--EIVTALKFHRASM--KEASE 128
Q QQ QQ QQH++Q++ + H + Q++ + +V+ + SM ++
Sbjct: 696 QHQQPQQQQQQHQQQLQLQQHMQQQLQQQQQQQETTSQLQSVVSPPQVGSPSMGVPPLNQ 875
Query: 129 QQQQQQLQQQQQQRAPDSPQLSQNPSFDQDGRYKSRRNPRIYPSCNTNFSSYMGDFSS 186
Q QQQ QQ QR P SPQ+S S NP P+ S +G SS
Sbjct: 876 QTQQQASPQQMSQRTPMSPQMSSGAI-----HAMSAGNPEACPASPQLSSQTLGSVSS 1034
Score = 27.7 bits (60), Expect = 9.0
Identities = 15/32 (46%), Positives = 17/32 (52%)
Frame = +3
Query: 121 ASMKEASEQQQQQQLQQQQQQRAPDSPQLSQN 152
A M QQQQ Q QQQQQ+ QL Q+
Sbjct: 663 AGMNLYMSQQQQHQQPQQQQQQHQQQLQLQQH 758
>TC206320 similar to UP|Q9W4F9 (Q9W4F9) CG32772-PA, partial (10%)
Length = 1120
Score = 43.1 bits (100), Expect = 2e-04
Identities = 34/131 (25%), Positives = 56/131 (41%), Gaps = 3/131 (2%)
Frame = +2
Query: 59 GQNLTKHGKIGKACQSQQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREIVTA--- 115
G NL + + Q Q QQ QQH++Q++ + H + Q++ ++ + V +
Sbjct: 320 GMNLYMSQQQQQQHQQPQPQQQQQQHQQQLQLQQHMQQQLQQQQQQETTSQLQAVVSPPQ 499
Query: 116 LKFHRASMKEASEQQQQQQLQQQQQQRAPDSPQLSQNPSFDQDGRYKSRRNPRIYPSCNT 175
+ + ++Q QQQ QQ QR P SPQ+S + NP P+
Sbjct: 500 VGSPSMGIPPMNQQAQQQASPQQMSQRTPMSPQMSSGAIHAMNA-----GNPEACPASPQ 664
Query: 176 NFSSYMGDFSS 186
S +G SS
Sbjct: 665 LSSQTLGSVSS 697
>CO036556
Length = 545
Score = 40.4 bits (93), Expect = 0.001
Identities = 33/108 (30%), Positives = 48/108 (43%), Gaps = 1/108 (0%)
Frame = +1
Query: 75 QQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREIVTALKFHRASMKEASEQQQQQQ 134
QQ QQ Q ++Q ++ LH SR + A ++ L+ R + +QQQQQ
Sbjct: 7 QQQQQQQQHQQQQQQQLLHMSRQSSQA------AAAQMNHLLQQQRLLQYQQHQQQQQQL 168
Query: 135 LQQQQQQRAPDSPQLSQNPSFDQDGRYKSRRNPRIYPS-CNTNFSSYM 181
L+ QQR+P S Q Q Q+ +S P P C + YM
Sbjct: 169 LKTMPQQRSPLSQQFQQ-----QNMPMRSPVKPAYEPGMCTRRLTHYM 297
>CO985160
Length = 777
Score = 40.4 bits (93), Expect = 0.001
Identities = 24/85 (28%), Positives = 40/85 (46%)
Frame = -2
Query: 73 QSQQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREIVTALKFHRASMKEASEQQQQ 132
Q Q Q QQH Q++++ Q++L + + ++++ + + + +QQQ
Sbjct: 740 QQHQQMQNQQQHLPQLQQQQQLPHMQQQQLPQLQQQQQQLPQLQQQQQQLPQLQQQQQQL 561
Query: 133 QQLQQQQQQRAPDSPQLSQNPSFDQ 157
QLQQQQQQ Q Q P Q
Sbjct: 560 PQLQQQQQQLPQLQQQQQQLPQLQQ 486
Score = 38.9 bits (89), Expect = 0.004
Identities = 23/78 (29%), Positives = 40/78 (50%)
Frame = -2
Query: 74 SQQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREIVTALKFHRASMKEASEQQQQQ 133
SQQ Q QQH++ ++ H + Q++ L +++ + L+ + + + +QQQQ
Sbjct: 764 SQQQQQMQQQHQQMQNQQQHLPQLQQQQQL--PHMQQQQLPQLQQQQQQLPQLQQQQQQL 591
Query: 134 QLQQQQQQRAPDSPQLSQ 151
QQQQQ+ P Q Q
Sbjct: 590 PQLQQQQQQLPQLQQQQQ 537
Score = 37.0 bits (84), Expect = 0.015
Identities = 26/88 (29%), Positives = 43/88 (48%), Gaps = 3/88 (3%)
Frame = -2
Query: 73 QSQQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREIVTALKFHRASMKEASEQQQQ 132
Q QQ Q QQ ++Q+ + Q++ L + +++ + L+ + + + +QQQQ
Sbjct: 638 QQQQQLPQLQQQQQQLPQLQQ-----QQQQLPQLQQQQQQLPQLQQQQQQLPQLQQQQQQ 474
Query: 133 ---QQLQQQQQQRAPDSPQLSQNPSFDQ 157
QQL Q QQQ+ P Q Q P Q
Sbjct: 473 VQQQQLAQLQQQQLPQIQQQQQLPQLQQ 390
Score = 32.7 bits (73), Expect = 0.28
Identities = 24/95 (25%), Positives = 43/95 (45%), Gaps = 1/95 (1%)
Frame = -2
Query: 58 HGQNLTKHGKIGKACQSQQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREIVTALK 117
H Q + + + Q QQ QQ Q++++ Q++ + + +++ +
Sbjct: 734 HQQMQNQQQHLPQLQQQQQLPHMQQQQLPQLQQQQQQLPQLQQQQQQLPQLQQQQQQLPQ 555
Query: 118 FHRASMKEASEQQQQQQLQQ-QQQQRAPDSPQLSQ 151
+ + QQQQQQL Q QQQQ+ QL+Q
Sbjct: 554 LQQQQQQLPQLQQQQQQLPQLQQQQQQVQQQQLAQ 450
Score = 30.4 bits (67), Expect = 1.4
Identities = 22/91 (24%), Positives = 42/91 (45%), Gaps = 6/91 (6%)
Frame = -2
Query: 73 QSQQSTQQPQQHKKQV------RRRLHTSRPYQERLLNMAEARREIVTALKFHRASMKEA 126
Q QQ Q QQ ++Q+ +++L + Q++L + + ++++ + + +
Sbjct: 608 QQQQQLPQLQQQQQQLPQLQQQQQQLPQLQQQQQQLPQLQQQQQQV------QQQQLAQL 447
Query: 127 SEQQQQQQLQQQQQQRAPDSPQLSQNPSFDQ 157
+QQ Q QQQQ + PQL Q Q
Sbjct: 446 QQQQLPQIQQQQQLPQLQQLPQLQQQQQLPQ 354
Score = 29.3 bits (64), Expect = 3.1
Identities = 14/36 (38%), Positives = 17/36 (46%)
Frame = -2
Query: 122 SMKEASEQQQQQQLQQQQQQRAPDSPQLSQNPSFDQ 157
S ++ + QQQ Q Q QQQ P Q Q P Q
Sbjct: 767 SSQQQQQMQQQHQQMQNQQQHLPQLQQQQQLPHMQQ 660
Score = 28.9 bits (63), Expect = 4.0
Identities = 18/68 (26%), Positives = 33/68 (48%)
Frame = -2
Query: 73 QSQQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREIVTALKFHRASMKEASEQQQQ 132
Q QQ Q QQ ++Q+ + + Q++ L + ++ + +++ + QQQ
Sbjct: 548 QQQQQLPQLQQQQQQLPQLQQQQQQVQQQQLAQLQQQQLPQIQQQQQLPQLQQLPQLQQQ 369
Query: 133 QQLQQQQQ 140
QQL Q QQ
Sbjct: 368 QQLPQLQQ 345
Score = 27.7 bits (60), Expect = 9.0
Identities = 33/123 (26%), Positives = 50/123 (39%), Gaps = 2/123 (1%)
Frame = -3
Query: 165 RNPRIYPS--CNTNFSSYMGDFSSYPPAFVPNSYPVPVASPIAPPPLMAENPNFILPNQP 222
RN R+ S C+ N S + S + +SY L N NF+ N
Sbjct: 775 RNCRVSSSNKCSNNISRCKTNSSIFRSCNNSSSY------------LTCNNSNFLSSNNS 632
Query: 223 LGLNLNFHDFNNLDATIHLSNTSLSSNSFSSATSSSQEVPSVELSQGDGISSLVNSTESN 282
NF ++ + SN S SN+F S +SSS S S + +S +S S+
Sbjct: 631 SN---NFLSCSSSSSNFLSSNNS--SNNFLSCSSSSNNFLSCSSSSNNFLSCNSSSNRSS 467
Query: 283 AAS 285
++S
Sbjct: 466 SSS 458
>TC213885 weakly similar to UP|Q9VUB7 (Q9VUB7) CG32133-PA, partial (4%)
Length = 540
Score = 40.0 bits (92), Expect = 0.002
Identities = 25/81 (30%), Positives = 43/81 (52%)
Frame = +2
Query: 77 STQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREIVTALKFHRASMKEASEQQQQQQLQ 136
S+QQ QQH++ ++ H + Q++ L +++ + L+ + + + +QQQ QLQ
Sbjct: 2 SSQQQQQHQQMQNQQQHLPQLQQQQQL--PHMQQQQLPQLQ-QQQQLPQLQQQQQLPQLQ 172
Query: 137 QQQQQRAPDSPQLSQNPSFDQ 157
QQQQ + PQL Q Q
Sbjct: 173 QQQQLQQQQLPQLQQQQQLPQ 235
Score = 38.5 bits (88), Expect = 0.005
Identities = 25/85 (29%), Positives = 42/85 (49%)
Frame = +2
Query: 73 QSQQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREIVTALKFHRASMKEASEQQQQ 132
Q Q Q QQH Q++++ Q++L + + ++++ + + + +Q QQ
Sbjct: 17 QQHQQMQNQQQHLPQLQQQQQLPHMQQQQLPQLQQ-QQQLPQLQQQQQLPQLQQQQQLQQ 193
Query: 133 QQLQQQQQQRAPDSPQLSQNPSFDQ 157
QQL Q QQQ+ PQL Q P Q
Sbjct: 194 QQLPQLQQQQ--QLPQLQQLPQLQQ 262
Score = 30.8 bits (68), Expect = 1.1
Identities = 19/69 (27%), Positives = 36/69 (51%)
Frame = +2
Query: 73 QSQQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREIVTALKFHRASMKEASEQQQQ 132
Q QQ Q PQ ++Q +L + Q++ L + ++++ + + +++ + Q
Sbjct: 110 QLQQQQQLPQLQQQQQLPQLQQQQQLQQQQLPQLQQQQQLPQLQQLPQ--LQQQQQLPQL 283
Query: 133 QQLQQQQQQ 141
QQLQ QQQQ
Sbjct: 284 QQLQPQQQQ 310
Score = 30.0 bits (66), Expect = 1.8
Identities = 23/94 (24%), Positives = 41/94 (43%)
Frame = +2
Query: 58 HGQNLTKHGKIGKACQSQQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREIVTALK 117
H Q + + + Q QQ QQ Q++++ + Q++ L + ++++
Sbjct: 23 HQQMQNQQQHLPQLQQQQQLPHMQQQQLPQLQQQQQLPQLQQQQQLPQLQQQQQLQQQQL 202
Query: 118 FHRASMKEASEQQQQQQLQQQQQQRAPDSPQLSQ 151
++ + QQ QLQQQQQ PQL Q
Sbjct: 203 PQLQQQQQLPQLQQLPQLQQQQQ-----LPQLQQ 289
>CA852848
Length = 329
Score = 37.4 bits (85), Expect = 0.011
Identities = 14/19 (73%), Positives = 18/19 (94%)
Frame = +1
Query: 363 NESCLDQCSVEYFQNSTLP 381
N+SCL+QCSV+YFQ S+LP
Sbjct: 16 NDSCLEQCSVDYFQESSLP 72
>TC213991 similar to UP|Q6H976 (Q6H976) STYLOSA protein, partial (12%)
Length = 440
Score = 37.0 bits (84), Expect = 0.015
Identities = 17/34 (50%), Positives = 21/34 (61%)
Frame = +3
Query: 124 KEASEQQQQQQLQQQQQQRAPDSPQLSQNPSFDQ 157
++ +QQQQQQ QQQQQQ+ P S Q P Q
Sbjct: 15 QQQQQQQQQQQQQQQQQQQQPQSQQQQSQPQQQQ 116
Score = 37.0 bits (84), Expect = 0.015
Identities = 18/41 (43%), Positives = 24/41 (57%)
Frame = +3
Query: 124 KEASEQQQQQQLQQQQQQRAPDSPQLSQNPSFDQDGRYKSR 164
++ +QQQQQQ QQQQQQ+ PQ Q S Q + + R
Sbjct: 6 QQQQQQQQQQQQQQQQQQQQQQQPQSQQQQSQPQQQQSRDR 128
Score = 33.5 bits (75), Expect = 0.16
Identities = 17/40 (42%), Positives = 20/40 (49%)
Frame = +3
Query: 129 QQQQQQLQQQQQQRAPDSPQLSQNPSFDQDGRYKSRRNPR 168
QQQQQQ QQQQQQ+ Q Q P Q ++ R
Sbjct: 3 QQQQQQQQQQQQQQQQQQQQQQQQPQSQQQQSQPQQQQSR 122
Score = 32.0 bits (71), Expect = 0.48
Identities = 16/39 (41%), Positives = 23/39 (58%)
Frame = +3
Query: 128 EQQQQQQLQQQQQQRAPDSPQLSQNPSFDQDGRYKSRRN 166
+QQQQQQ QQQQQQ+ Q Q S Q + + +++
Sbjct: 3 QQQQQQQQQQQQQQQQQQQQQQQQPQSQQQQSQPQQQQS 119
>BQ253857
Length = 422
Score = 36.2 bits (82), Expect = 0.025
Identities = 34/120 (28%), Positives = 56/120 (46%)
Frame = +3
Query: 25 EEEPPFSGAYMRSLVKQLRTNDQGCVVNSDASSHGQNLTKHGKIGKACQSQQSTQQPQQH 84
+ P S S++K+ R ++ NS +SS + T K+ + ++ T++P
Sbjct: 75 KNSPSSSDIQTTSIMKRTRKPNRRYYHNSSSSSSSSSSTTSFKLHRR-GNRTKTRKP--- 242
Query: 85 KKQVRRRLHTSRPYQERLLNMAEARREIVTALKFHRASMKEASEQQQQQQLQQQQQQRAP 144
K V +L S P ++ MA + + +A + QQQQQLQQQQQQ+ P
Sbjct: 243 -KFVSLKLQLSEPKKK----MASTKPK----------PKPKAEQLQQQQQLQQQQQQQQP 377
>BU965513
Length = 452
Score = 35.4 bits (80), Expect = 0.043
Identities = 29/96 (30%), Positives = 45/96 (46%), Gaps = 8/96 (8%)
Frame = +2
Query: 57 SHGQNLTKHGKIGKACQSQ-QSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREIVTA 115
S G + T + QSQ QS Q P +Q +++ S+P+Q+ L + ++
Sbjct: 182 SSGGSWTMIPTHNSSIQSQSQSNQDPNLFLQQQQQQFLQSQPFQQALPPQSPFQQH---- 349
Query: 116 LKFHRASMKEASEQQQQ-------QQLQQQQQQRAP 144
H ++ +QQQQ QQ QQQQQQ+ P
Sbjct: 350 ---HHLYQQQQQQQQQQRLLPQPQQQQQQQQQQQQP 448
>TC226370 similar to UP|Q9M0H8 (Q9M0H8) Predicted proline-rich protein,
partial (43%)
Length = 1810
Score = 35.4 bits (80), Expect = 0.043
Identities = 28/83 (33%), Positives = 38/83 (45%), Gaps = 2/83 (2%)
Frame = +2
Query: 128 EQQQQQQLQQQQQQRAPDSPQL--SQNPSFDQDGRYKSRRNPRIYPSCNTNFSSYMGDFS 185
+QQQQQ QQQ QQ+ S Q+ SQ P R S P +YP N ++ +
Sbjct: 875 QQQQQQPQQQQPQQQQQWSQQVLPSQPPPMQSQVRPPS---PNVYPPYQPNQAT-----N 1030
Query: 186 SYPPAFVPNSYPVPVASPIAPPP 208
P +PNS + + PPP
Sbjct: 1031PSPAETLPNSMAMQMPYSGVPPP 1099
>TC231181 weakly similar to UP|Q86PD9 (Q86PD9) RE65015p, partial (10%)
Length = 671
Score = 35.4 bits (80), Expect = 0.043
Identities = 26/94 (27%), Positives = 46/94 (48%), Gaps = 1/94 (1%)
Frame = +2
Query: 62 LTKHGKIGKACQSQQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREIVTALKFHRA 121
+TK+ ++ + Q QQ Q QQH Q++++ Q++L + ++ L+ +
Sbjct: 221 MTKNNQLWQYYQQQQQQLQLQQHYIQLQQQSFQQGLSQQQL------QHSLLEPLQPQQL 382
Query: 122 SMKEASEQQQQQ-QLQQQQQQRAPDSPQLSQNPS 154
+ +QQQQ Q QQ QQ P++ Q PS
Sbjct: 383 QQQVLQQQQQQYFQQQQPLQQEHPENLMQQQQPS 484
>TC226371 similar to UP|Q9M0H8 (Q9M0H8) Predicted proline-rich protein,
partial (10%)
Length = 1099
Score = 35.0 bits (79), Expect = 0.056
Identities = 26/80 (32%), Positives = 35/80 (43%)
Frame = +1
Query: 128 EQQQQQQLQQQQQQRAPDSPQLSQNPSFDQDGRYKSRRNPRIYPSCNTNFSSYMGDFSSY 187
+ QQQQQ QQQQQ+ Q SQ P R S P +YP N ++ +
Sbjct: 181 QPQQQQQPPQQQQQQWSQQVQPSQPPPMQSQVRPSS---PNVYPPYQPNQAT-----NPS 336
Query: 188 PPAFVPNSYPVPVASPIAPP 207
P +PNS + + PP
Sbjct: 337 PAETLPNSMAMQMPYSGVPP 396
>TC206957 weakly similar to UP|Q9LUI1 (Q9LUI1) Extensin protein-like, partial
(7%)
Length = 595
Score = 34.7 bits (78), Expect = 0.074
Identities = 26/62 (41%), Positives = 29/62 (45%)
Frame = +2
Query: 161 YKSRRNPRIYPSCNTNFSSYMGDFSSYPPAFVPNSYPVPVASPIAPPPLMAENPNFILPN 220
Y S +P YP SSY SSYPP P+SYP A P PP A P+ I P
Sbjct: 245 YSSHSSP--YPP-----SSYPPQPSSYPPPPPPSSYPPASAYPYPPP---AGYPSGIYPP 394
Query: 221 QP 222
P
Sbjct: 395 PP 400
>BE807600 similar to GP|24461867|gb CTV.22 {Poncirus trifoliata}, partial
(4%)
Length = 414
Score = 34.3 bits (77), Expect = 0.096
Identities = 22/67 (32%), Positives = 31/67 (45%)
Frame = +3
Query: 75 QQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREIVTALKFHRASMKEASEQQQQQQ 134
QQ Q QQ + Q ++ + Q +LL K + ++ +QQQQQQ
Sbjct: 237 QQVVPQQQQQQSQNSQQYIYQQQMQHQLLRQ-----------KLQQQQQQQQQQQQQQQQ 383
Query: 135 LQQQQQQ 141
QQQQQQ
Sbjct: 384 QQQQQQQ 404
Score = 32.0 bits (71), Expect = 0.48
Identities = 23/91 (25%), Positives = 40/91 (43%), Gaps = 2/91 (2%)
Frame = +3
Query: 60 QNLTKHGKIGKACQSQQSTQQ--PQQHKKQVRRRLHTSRPYQERLLNMAEARREIVTALK 117
QN+ +G + Q P ++ R+ + Q++ N +++ I
Sbjct: 138 QNIPGQNSVGSTISQNSNMQNMFPGSQRQIQGRQQVVPQQQQQQSQN---SQQYIYQQQM 308
Query: 118 FHRASMKEASEQQQQQQLQQQQQQRAPDSPQ 148
H+ ++ +QQQQQQ QQQQQQ+ Q
Sbjct: 309 QHQLLRQKLQQQQQQQQQQQQQQQQQQQQQQ 401
Score = 31.6 bits (70), Expect = 0.62
Identities = 15/24 (62%), Positives = 16/24 (66%)
Frame = +3
Query: 129 QQQQQQLQQQQQQRAPDSPQLSQN 152
QQQQQQ QQQQQQ+ Q QN
Sbjct: 336 QQQQQQQQQQQQQQQQQQQQQQQN 407
Score = 31.2 bits (69), Expect = 0.81
Identities = 23/91 (25%), Positives = 41/91 (44%), Gaps = 2/91 (2%)
Frame = +3
Query: 52 NSDASSHGQNLTKHGKIGKACQSQQSTQQ--PQQHKKQVRRRLHTSRPYQERLLNMAEAR 109
NS S+ QN + + Q QQ PQQ ++Q + ++ + + +
Sbjct: 156 NSVGSTISQNSNMQNMFPGSQRQIQGRQQVVPQQQQQQSQN--------SQQYIYQQQMQ 311
Query: 110 REIVTALKFHRASMKEASEQQQQQQLQQQQQ 140
+++ + ++ +QQQQQQ QQQQQ
Sbjct: 312 HQLLRQKLQQQQQQQQQQQQQQQQQQQQQQQ 404
>TC217981 similar to UP|Q9VSK3 (Q9VSK3) CG7015-PA, partial (3%)
Length = 738
Score = 33.9 bits (76), Expect = 0.13
Identities = 15/21 (71%), Positives = 17/21 (80%)
Frame = +3
Query: 129 QQQQQQLQQQQQQRAPDSPQL 149
QQQ+QQ QQQQQQ+ P PQL
Sbjct: 153 QQQEQQQQQQQQQQHPPQPQL 215
Score = 27.7 bits (60), Expect = 9.0
Identities = 13/37 (35%), Positives = 18/37 (48%)
Frame = +3
Query: 112 IVTALKFHRASMKEASEQQQQQQLQQQQQQRAPDSPQ 148
+ T + + Q QQQ+ QQQQQQ+ PQ
Sbjct: 96 VATGAGLQQQQHHQHHHQHQQQEQQQQQQQQQQHPPQ 206
>TC228352 weakly similar to GB|AAP13389.1|30023712|BT006281 At1g55080
{Arabidopsis thaliana;} , partial (50%)
Length = 1007
Score = 33.5 bits (75), Expect = 0.16
Identities = 27/97 (27%), Positives = 41/97 (41%)
Frame = +3
Query: 50 VVNSDASSHGQNLTKHGKIGKACQSQQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEAR 109
+V S+ +G + I QS Q P +Q ++ L S+P+Q+ L
Sbjct: 129 IVESEMDHYGSSGGSWTMIPTHNSQSQSNQDPNLFLQQQQQFLQ-SQPFQQAL------- 284
Query: 110 REIVTALKFHRASMKEASEQQQQQQLQQQQQQRAPDS 146
A +QQQQ+ LQQQQQQ+ P +
Sbjct: 285 -----------APQSPFQQQQQQRLLQQQQQQQQPQN 362
>TC220413 similar to UP|Q9LF51 (Q9LF51) Glutamine-rich protein, partial (31%)
Length = 761
Score = 32.7 bits (73), Expect = 0.28
Identities = 32/100 (32%), Positives = 47/100 (47%), Gaps = 4/100 (4%)
Frame = +1
Query: 120 RASMKEASEQQQQQQ--LQQQQQQRAPDSPQLSQNPSFDQDGRYKSRRNPRIYPSCNTNF 177
+A ++ +QQQQQQ LQQQQQQ+ L Q Q + ++ R +PS N
Sbjct: 265 KALHQQQQQQQQQQQLLLQQQQQQQHQQHYLLLQQLQKQQQQQQQAAAISR-FPS---NI 432
Query: 178 SSYMGDFSSYPPAFVPNSYPVPVASPIAPPPL--MAENPN 215
+++ PN P P +P P PL + +NPN
Sbjct: 433 DAHLRPIRPLNLQQNPNPSPNPAPNP-NPNPLINLHQNPN 549
>TC207343 similar to GB|AAN31116.1|23506209|AY149962 At3g06130/F28L1_7
{Arabidopsis thaliana;} , partial (14%)
Length = 540
Score = 32.7 bits (73), Expect = 0.28
Identities = 23/66 (34%), Positives = 33/66 (49%)
Frame = +1
Query: 124 KEASEQQQQQQLQQQQQQRAPDSPQLSQNPSFDQDGRYKSRRNPRIYPSCNTNFSSYMGD 183
++ +QQQQQQLQQQQQ A ++Q + D R++ R P+ N + Y
Sbjct: 43 QQQQQQQQQQQLQQQQQYMA---AMMNQQRAIGND-RFQPMMYARPPPAVNYMYPPY--- 201
Query: 184 FSSYPP 189
YPP
Sbjct: 202 --PYPP 213
>TC227037 homologue to UP|MSI4_ARATH (O22607) WD-40 repeat protein MSI4,
partial (86%)
Length = 1945
Score = 32.7 bits (73), Expect = 0.28
Identities = 14/33 (42%), Positives = 19/33 (57%)
Frame = +2
Query: 119 HRASMKEASEQQQQQQLQQQQQQRAPDSPQLSQ 151
H +KE + QQQQQ QQQQ+ D P + +
Sbjct: 17 HGKGLKEGRKTQQQQQHHHQQQQQQQDQPSVDE 115
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.312 0.126 0.365
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,080,676
Number of Sequences: 63676
Number of extensions: 272922
Number of successful extensions: 3429
Number of sequences better than 10.0: 164
Number of HSP's better than 10.0 without gapping: 2262
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2846
length of query: 382
length of database: 12,639,632
effective HSP length: 99
effective length of query: 283
effective length of database: 6,335,708
effective search space: 1793005364
effective search space used: 1793005364
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0133.10


