

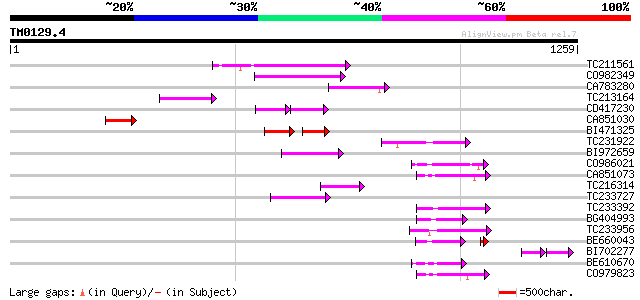
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0129.4
(1259 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC211561 weakly similar to UP|O22675 (O22675) Reverse transcript... 116 7e-26
CO982349 84 4e-16
CA783280 weakly similar to PIR|T01871|T01 RNA-directed DNA polym... 79 1e-14
TC213164 78 3e-14
CD417230 weakly similar to GP|2244915|emb| reverse transcriptase... 52 9e-14
CA851030 76 1e-13
BI471325 58 2e-13
TC231922 73 9e-13
BI972659 72 2e-12
CO986021 69 1e-11
CA851073 69 2e-11
TC216314 similar to GB|AAP68269.1|31711826|BT008830 At1g70320 {A... 67 5e-11
TC233727 similar to UP|Q9FKH8 (Q9FKH8) Similarity to non-LTR ret... 66 1e-10
TC233392 64 4e-10
BG404993 64 4e-10
TC233956 63 7e-10
BE660043 60 1e-09
BI702277 47 1e-09
BE610670 61 3e-09
CO979823 61 4e-09
>TC211561 weakly similar to UP|O22675 (O22675) Reverse transcriptase
(Fragment) , partial (59%)
Length = 1077
Score = 116 bits (290), Expect = 7e-26
Identities = 89/312 (28%), Positives = 139/312 (44%), Gaps = 6/312 (1%)
Frame = +1
Query: 451 IDNALVVYECFHFMKKRISGRNGMMTLKLDMSKAYDRVEWSFLQGVLQKMGFPPSWVSLI 510
+ +AL+ E K+ + K+D KAYD V W+FL ++ +M F P W+ I
Sbjct: 1 LHSALIANEVIDEAKR---SNKSCLVFKVDYEKAYDSVSWNFLMYMMWRMDFSPKWIKWI 171
Query: 511 ------MSVSPLLGFPLCLTEIPSRVLLLLEAYVRVILFRLIFLSYVERCSQLLLRSVWL 564
S+S L+ +P R L + L +F E + L+ R+V
Sbjct: 172 EECVKSASISVLVNGSPTAEFLPQRGLRQGDP-----LTPFLFNIVAEGLNGLMRRAVEE 336
Query: 565 LMLHGIKVANRAPVISHLLFADDSIIFARANTQEAECVLNILSTYERASGQVINLDKSQL 624
+ V IS L +ADD+I F A + E + IL ++E S IN KS
Sbjct: 337 NLYKPYLVGANGVPISILQYADDTIFFGEAAKENVEAIKVILRSFELVSNLRINFAKSCF 516
Query: 625 SVSRNVPQNGFNELKQLLNVNAVESFDKYLGLPTMIGKFKTRIFNFVKDRVWKKLKGWKE 684
V V E L+ + + YLG+P + +++ + + +KL WK
Sbjct: 517 GVF-GVTDQWKQEAANYLHCSLLAFPFIYLGIPIGANSRRCHMWDSIIRKCERKLSKWK* 693
Query: 685 STLSRAGREVLIKSVVQAIPSYVMSCFILPDSLCADIERMVSRFYWGGDVDKRGLQWASW 744
+S GR LIKSV+ +IP Y S F +P S+ + ++ F WGG D++ + W W
Sbjct: 694 RHISFGGRVTLIKSVLTSIPIYFFSFFRVPQSVVDKLVKI*RTFLWGGGPDQKKISWIRW 873
Query: 745 KKLTRSKFDGGL 756
+KL K GG+
Sbjct: 874 EKLCLPKERGGI 909
>CO982349
Length = 795
Score = 84.0 bits (206), Expect = 4e-16
Identities = 54/201 (26%), Positives = 89/201 (43%)
Frame = +3
Query: 545 LIFLSYVERCSQLLLRSVWLLMLHGIKVANRAPVISHLLFADDSIIFARANTQEAECVLN 604
L+F E + L+ +V + V +S L +ADD+I F A + + +
Sbjct: 180 LLFNIVAEGLTGLMREAVDRKRFNSFLVGKNKEPVSILQYADDTIFFEEATMENMKVIKT 359
Query: 605 ILSTYERASGQVINLDKSQLSVSRNVPQNGFNELKQLLNVNAVESFDKYLGLPTMIGKFK 664
IL +E ASG IN +S+ P + E + LN + + YLG+P
Sbjct: 360 ILRCFELASGLKINFAESRFGAIWK-PDHWCKEAAEFLNCSMLSMPFSYLGIPIEANPRC 536
Query: 665 TRIFNFVKDRVWKKLKGWKESTLSRAGREVLIKSVVQAIPSYVMSCFILPDSLCADIERM 724
I + + + KL WK+ +S GR LI +++ A+P Y S F P + + +
Sbjct: 537 REI*DPIIRKCETKLARWKQRHISLGGRVTLINAILTALPIYFFSFFRAPSKVVQKLVNI 716
Query: 725 VSRFYWGGDVDKRGLQWASWK 745
+F WGG +R + W W+
Sbjct: 717 QRKFLWGGGSXQRRIAWVKWE 779
>CA783280 weakly similar to PIR|T01871|T01 RNA-directed DNA polymerase
homolog T24M8.8 - Arabidopsis thaliana, partial (4%)
Length = 456
Score = 79.0 bits (193), Expect = 1e-14
Identities = 40/139 (28%), Positives = 66/139 (46%), Gaps = 5/139 (3%)
Frame = +2
Query: 709 SCFILPDSLCADIERMVSRFYWGGDVDKRGLQWASWKKLTRSKFDGGLGFRDFKSFNIAL 768
S F LP + A +E + RF WGG+ D R + W +WK + K GGLG +D ++FN L
Sbjct: 5 SFFSLP*KIIAKLESLQRRFLWGGEADSRKIAWVNWKTVCLPKAKGGLGIKDLRTFNTTL 184
Query: 769 VAKNWWRIYTQPETLLAQVFKGVYFAQGDLLGAKRGYRPSYAWSSILQSSWI-----FQE 823
+ K W ++ + A+V + Y L G + S W ++++ + +
Sbjct: 185 LGKWRWDLFYIQQEPWAKVLQSKYGGWRALEEGSSGSKDSAWWKDLIKTQQLQRNIPLKR 364
Query: 824 GGRWKIGDGSQVDILHDQW 842
WK+G G ++ D W
Sbjct: 365 ETIWKVGGGDRIKFWEDLW 421
>TC213164
Length = 446
Score = 77.8 bits (190), Expect = 3e-14
Identities = 40/128 (31%), Positives = 68/128 (52%)
Frame = +3
Query: 332 FTREEVEEALFQMHPTKAPGLDGFPTLFYQKYWDIIGDDVSAFCLQVLQGVIPPGMINQT 391
F ++EV ++ K+PG DG F +++WDI+ D+ F + + N
Sbjct: 30 FEKKEVRATMWDRGNAKSPGPDGLNFKFIKEFWDILKTDLLRFLDEFYANGVFLKRSNAF 209
Query: 392 LLVLIPKIMKPEHATQFRPISLCNVLFKIITKTIANRLKIILPDIICQTQSAFVPGRLII 451
L LIPK+ P +++PISL ++KI+ K ++ R K +LP II + Q+ F+ GR +
Sbjct: 210 FLALIPKVHDP*SLNEYKPISLIGCIYKIVAKLLSGRHKKVLPTIIDERQTVFMEGRHTL 389
Query: 452 DNALVVYE 459
N ++ E
Sbjct: 390 HNVVIANE 413
>CD417230 weakly similar to GP|2244915|emb| reverse transcriptase like
protein {Arabidopsis thaliana}, partial (7%)
Length = 688
Score = 52.4 bits (124), Expect(2) = 9e-14
Identities = 29/86 (33%), Positives = 46/86 (52%)
Frame = -2
Query: 623 QLSVSRNVPQNGFNELKQLLNVNAVESFDKYLGLPTMIGKFKTRIFNFVKDRVWKKLKGW 682
+ S S+NV ++ L + + + KYLG+P + F+F+ D+V K+ W
Sbjct: 333 KFSFSKNVNWWVEEDISSGLGLQSTDDLGKYLGVPIFHKNPTSHTFDFIIDKVNKRFSRW 154
Query: 683 KESTLSRAGREVLIKSVVQAIPSYVM 708
K + LS GR L K VV A+PS++M
Sbjct: 153 KSNLLSMVGRLTLTK*VV*ALPSHIM 76
Score = 43.9 bits (102), Expect(2) = 9e-14
Identities = 24/78 (30%), Positives = 42/78 (53%)
Frame = -3
Query: 546 IFLSYVERCSQLLLRSVWLLMLHGIKVANRAPVISHLLFADDSIIFARANTQEAECVLNI 605
+F+ +ER S L+ + + I+V +ISHL F DD I+F AN + + +
Sbjct: 566 LFVLCLERLSHLINLVMNKKL*RSIQVNRGGLLISHLAFVDDLILFVEANMDQVDVINLT 387
Query: 606 LSTYERASGQVINLDKSQ 623
L + +SG +N+DK++
Sbjct: 386 LELFSDSSGAKVNVDKTK 333
>CA851030
Length = 358
Score = 75.9 bits (185), Expect = 1e-13
Identities = 34/67 (50%), Positives = 46/67 (67%)
Frame = +1
Query: 214 QTKEVLMAINVAERDLDSLLEQEEVWWKQRSRASWLKHGDKNTRFFHQKANQRRKRNLIE 273
+ ++V+ + V E LDSLL+QEE +W+QRSRA W K DKNT FFH+K QR+ N I
Sbjct: 106 KNRQVIASTKVVEESLDSLLKQEEEYWQQRSRAIWFKERDKNTEFFHRKTLQRKASNSIT 285
Query: 274 VLKDDNG 280
++DD G
Sbjct: 286 KIQDDEG 306
>BI471325
Length = 421
Score = 57.8 bits (138), Expect(2) = 2e-13
Identities = 28/66 (42%), Positives = 42/66 (63%)
Frame = +2
Query: 567 LHGIKVANRAPVISHLLFADDSIIFARANTQEAECVLNILSTYERASGQVINLDKSQLSV 626
+H +KV ++SHLL D +F R +EA+ + NIL+T+E +SG V+NL KS++
Sbjct: 8 IHRVKVHRGITILSHLLSVDFYFLFCRTIDKEADVLKNILTTFEASSGLVVNLRKSEICF 187
Query: 627 SRNVPQ 632
SRN Q
Sbjct: 188 SRNTSQ 205
Score = 37.4 bits (85), Expect(2) = 2e-13
Identities = 23/62 (37%), Positives = 38/62 (61%), Gaps = 3/62 (4%)
Frame = +1
Query: 651 DKYLGLPTMIGKFKTRIFNFVKD---RVWKKLKGWKESTLSRAGREVLIKSVVQAIPSYV 707
D+YL +P+MI K IFN+++D R++ L +ES + + ++IK V Q+I +Y
Sbjct: 238 DEYLDMPSMIRTKKKAIFNYLRDEFGRIYSTLV-QQESFKGQREKCLIIKYVAQSITTYC 414
Query: 708 MS 709
MS
Sbjct: 415 MS 420
>TC231922
Length = 803
Score = 72.8 bits (177), Expect = 9e-13
Identities = 54/213 (25%), Positives = 91/213 (42%), Gaps = 16/213 (7%)
Frame = -3
Query: 827 WKIGDGSQVDILHDQWLPQGVPVIGSQDLMAEFG------VSKVSHLIDHAAK---SWKY 877
W++G G ++ D WL +G + + + +SK+ +A W+
Sbjct: 705 WRVGCGDKIKFWQDSWLSEGCNLQQKYNQLYTISRQ*NLTISKMGKFSQNARSWEFKWRR 526
Query: 878 ALVDFIFHPAT--VSHILKIHLPLNGSVDTLMWPETVDGNYCSKSGYIFVRKNFLGACSS 935
L D+ + A ++ I I + N + D++ W G Y +KS Y +
Sbjct: 525 RLFDYEYAMAVDFMNEISGISIQ-NQAHDSMFWKADSSGVYSTKSAYRLL---------- 379
Query: 936 TYSQPSL-PAP---LWKKFWRTSAQPRCKEVAWRVVSSLLPIRAALHRRGMDV-DPLCPV 990
PS+ PAP +++ W PR +WR+ LP R L RR + + D +CP+
Sbjct: 378 ---MPSISPAPSRRIFQILWHLKIPPRAAVFSWRLFLDRLPTRGNLSRRSIPIQDIMCPL 208
Query: 991 CANEEETVFHLFVSCPVAQNFWFGSPLSLRVHG 1023
C + E HLF C + + W+ S +RV G
Sbjct: 207 CGCQHEEAGHLFFHCKMTKGLWWESMRWIRVIG 109
>BI972659
Length = 453
Score = 72.0 bits (175), Expect = 2e-12
Identities = 42/138 (30%), Positives = 68/138 (48%)
Frame = +1
Query: 604 NILSTYERASGQVINLDKSQLSVSRNVPQNGFNELKQLLNVNAVESFDKYLGLPTMIGKF 663
++L +E SG IN KSQ + P N ++ QLLN ++ YLG+P +
Sbjct: 40 SMLRGFEMVSGLRINYAKSQFGIVGFQP-NWAHDAAQLLNCRQLDIPFHYLGMPIAVKAS 216
Query: 664 KTRIFNFVKDRVWKKLKGWKESTLSRAGREVLIKSVVQAIPSYVMSCFILPDSLCADIER 723
++ + ++ KL W + LS GR LIKSV+ A+P Y++S F +P + +
Sbjct: 217 SRMVWEPLINKFKAKLSKWNQKYLSMGGRVTLIKSVLNALPIYLLSFFKIPQRIVDKLVS 396
Query: 724 MVSRFYWGGDVDKRGLQW 741
+ F WGG+ + W
Sbjct: 397 LQRTFMWGGNQHHNRISW 450
>CO986021
Length = 900
Score = 69.3 bits (168), Expect = 1e-11
Identities = 49/180 (27%), Positives = 81/180 (44%), Gaps = 9/180 (5%)
Frame = -1
Query: 893 LKIHLPLNGSVDTLMWPETVDGNYCSKSGYIFVRKNFLGACSSTYSQPSLPAPLWKKFWR 952
+ IH L D+++W G Y +KS Y + N ++P + +K W+
Sbjct: 807 ISIHQQLQ---DSMLWKADPTGIYSTKSAYRLLLPN---------NRPGQHSSNFKILWK 664
Query: 953 TSAQPRCKEVAWRVVSSLLPIRAALHRRGMDV-DPLCPVCANEEETVFHLFVSCPVAQNF 1011
PR + +WR+ L IRA L RR + + D +CP+C N +E HLF C +
Sbjct: 663 LKIPPRAELFSWRLFRDRLSIRANLLRRHVALQDIMCPLCGNHQEEAGHLFFHCRMTIGL 484
Query: 1012 WFGSPLSLRVHGFSSMEDFLADFFRAAD--------DDALALWQAGVYALWEMRNRVVFR 1063
W+ S R G + ++D A F + +D + W A +W+ RN ++F+
Sbjct: 483 WWESMNWTRTLG-AFLDDPAAHFIQFSDGFGAQRNHNRRCLWWIALTSTIWQHRNSLLFK 307
>CA851073
Length = 633
Score = 68.6 bits (166), Expect = 2e-11
Identities = 46/171 (26%), Positives = 77/171 (44%), Gaps = 8/171 (4%)
Frame = +1
Query: 904 DTLMWPETVDGNYCSKSGYIFVRKNFLGACSSTYSQPSLPAPLWKKFWRTSAQPRCKEVA 963
DT++W G Y +KS Y L C+ S+ A ++K W+ PR +
Sbjct: 28 DTMVWKADPCGVYSTKSAY-----RILMTCNRHVSE----ANIFKTIWKLKIPPRAAVFS 180
Query: 964 WRVVSSLLPIRAALHRRGMDV-DPLCPVCANEEETVFHLFVSCPVAQNFWFGSPLSLRVH 1022
WR++ LP R L RR + + + CP+C E+E HLF +C + + W+ S ++
Sbjct: 181 WRLIKDRLPTRHNLLRRNVSIQENECPLCGYEQEEADHLFFNCKMTRGLWWESMRWNQIV 360
Query: 1023 GFSSMEDF-----LADFFRAADDDA--LALWQAGVYALWEMRNRVVFRDGE 1066
G S+ D F A + W A ++W+ RN ++F+ +
Sbjct: 361 GPLSVSPASHFVQFCDGFGAGRNHTRWCGWWIALTSSIWKHRNLLIFQGNQ 513
>TC216314 similar to GB|AAP68269.1|31711826|BT008830 At1g70320 {Arabidopsis
thaliana;} , partial (18%)
Length = 759
Score = 67.0 bits (162), Expect = 5e-11
Identities = 32/97 (32%), Positives = 49/97 (49%)
Frame = +2
Query: 691 GREVLIKSVVQAIPSYVMSCFILPDSLCADIERMVSRFYWGGDVDKRGLQWASWKKLTRS 750
GR LIKSV+ A+P +S F +P + + + +F WGG + W W +
Sbjct: 8 GRVTLIKSVLSALPIXXLSFFKIPQRIVDKLVTLQRQFLWGGTQHHNRIPWVKWADICNP 187
Query: 751 KFDGGLGFRDFKSFNIALVAKNWWRIYTQPETLLAQV 787
K DGGLG +D +FN AL + W + + L A++
Sbjct: 188 KIDGGLGIKDLSNFNAALRGRWIWGLASNHNQLWARL 298
>TC233727 similar to UP|Q9FKH8 (Q9FKH8) Similarity to non-LTR retroelement
reverse transcriptase, partial (7%)
Length = 956
Score = 65.9 bits (159), Expect = 1e-10
Identities = 40/132 (30%), Positives = 65/132 (48%)
Frame = -2
Query: 580 SHLLFADDSIIFARANTQEAECVLNILSTYERASGQVINLDKSQLSVSRNVPQNGFNELK 639
SH++ ADD +IF + ++ ++N Y+ + GQVI+ KS++ + ++P +
Sbjct: 451 SHVMHADDIMIFCKGTSRNVHNLINFFQLYKDSFGQVISKHKSKIFIG-SIPGQRLRVIS 275
Query: 640 QLLNVNAVESFDKYLGLPTMIGKFKTRIFNFVKDRVWKKLKGWKESTLSRAGREVLIKSV 699
LL + Y G+P GK R V D++ KL WK S LS GR L+ SV
Sbjct: 274 DLLQYFHSDIPFNYFGVPIFKGKPTRRHLYCVADKIKCKLASWKGSILSIMGRIQLVNSV 95
Query: 700 VQAIPSYVMSCF 711
+ + Y S +
Sbjct: 94 IHGMLLYSFSVY 59
Score = 37.0 bits (84), Expect = 0.055
Identities = 21/65 (32%), Positives = 36/65 (55%), Gaps = 3/65 (4%)
Frame = -3
Query: 450 IIDNALVVYECFHFMKKRISGRNGMMTLKLDMSKAYDRVEWSFLQGVLQKMGFPP---SW 506
I D V E + + K++ G N + LK+D+ KA+D ++ FL VL K G+ +W
Sbjct: 834 IKDCTCVTSEVINMLDKKVFGGN--LALKIDIVKAFDTLDRDFLISVLNKFGYSHTFCNW 661
Query: 507 VSLIM 511
+ +I+
Sbjct: 660 IRVIL 646
>TC233392
Length = 1145
Score = 63.9 bits (154), Expect = 4e-10
Identities = 43/171 (25%), Positives = 73/171 (42%), Gaps = 8/171 (4%)
Frame = -1
Query: 904 DTLMWPETVDGNYCSKSGYIFVRKNFLGACSSTYSQPSLPAPLWKKFWRTSAQPRCKEVA 963
DT +W + Y ++S Y ++ + G Q W+ + A
Sbjct: 656 DTWVWKHEPNELYSTRSAYKLLQGHLEGEDQDGALQD---------LWKLKIPAKASIFA 504
Query: 964 WRVVSSLLPIRAALHRRGMDV-DPLCPVCANEEETVFHLFVSCPVAQNFWFGSPL---SL 1019
WR++ LP ++ LHRR + + D LCP C EE H+F+ C + W+ S SL
Sbjct: 503 WRLIRDRLPTKSNLHRRQVVLEDSLCPFCRIREEDASHIFLECNKIRPLWWESQTWVRSL 324
Query: 1020 RVHGFSSMEDFLADFFRAADDDALALWQ----AGVYALWEMRNRVVFRDGE 1066
V + + FL W+ A ++LW+ RN+V+F++ +
Sbjct: 323 GVSPINKRQHFLQHVHGTPGSKRYNRWKTWWIALTWSLWKHRNQVIFQNAQ 171
>BG404993
Length = 412
Score = 63.9 bits (154), Expect = 4e-10
Identities = 32/113 (28%), Positives = 53/113 (46%), Gaps = 1/113 (0%)
Frame = -3
Query: 904 DTLMWPETVDGNYCSKSGYIFVRKNFLGACSSTYSQPSLPAPLWKKFWRTSAQPRCKEVA 963
D+ W GNY +K+ Y +++ G ++ + W+ P+
Sbjct: 332 DSWTWKADPSGNYSTKTAYELLQEAIRGDNEDG---------IFVQLWKLKIPPKAPVFT 180
Query: 964 WRVVSSLLPIRAALHRRGMDV-DPLCPVCANEEETVFHLFVSCPVAQNFWFGS 1015
WR+++ LP + L RR +++ D LCP+C N EE HLF +C FW+ S
Sbjct: 179 WRLINDRLPTKVNLRRRQVEISDSLCPLCNNSEEDAAHLFFNCNKTLPFWWES 21
>TC233956
Length = 757
Score = 63.2 bits (152), Expect = 7e-10
Identities = 49/195 (25%), Positives = 81/195 (41%), Gaps = 14/195 (7%)
Frame = +2
Query: 889 VSHILKIHLPLNGSVDTLMWPETVDGNYCSKSGYIFVRKNF------LGACSSTYSQPSL 942
+ HI +I L N + DT +W G +KS Y ++ LG
Sbjct: 17 IDHIHEIRLNNNFN-DTWVWRAESTGIISTKSAYQVIKSELDDEGQHLG----------- 160
Query: 943 PAPLWKKFWRTSAQPRCKEVAWRVVSSLLPIRAALHRRGMDV-DPLCPVCANEEETVFHL 1001
+KK W P+ WR++ LP + L +R + V D LCP C ++ ET HL
Sbjct: 161 ----FKKLWEIKVPPKALSFVWRLLWDRLPTKDNLIKRQIQVEDDLCPFCHSQSETASHL 328
Query: 1002 FVSCPVAQNFWF---GSPLSLRVHGFSSMEDFLADFFRAA----DDDALALWQAGVYALW 1054
F +C W+ +V M++FL + AA + W A ++W
Sbjct: 329 FFTCGKIMPLWWEFLSWVKEDKVFHCRPMDNFLQHYSSAASKVSNTRRTMWWIAVTNSIW 508
Query: 1055 EMRNRVVFRDGEIPV 1069
+RN ++F++ + +
Sbjct: 509 RLRNDIIFQNQTVDI 553
>BE660043
Length = 714
Score = 59.7 bits (143), Expect(2) = 1e-09
Identities = 31/111 (27%), Positives = 55/111 (48%), Gaps = 1/111 (0%)
Frame = -2
Query: 902 SVDTLMWPETVDGNYCSKSGYIFVRKNFLGACSSTYSQPSLPAPLWKKFWRTSAQPRCKE 961
+ D+ +W +G Y S++ YI ++ N S+ ++K W+ +
Sbjct: 632 AADSWVWKPESNGYYSSRNAYILLQGN---------SEEGNMDDIFKDLWKLKIPAKASI 480
Query: 962 VAWRVVSSLLPIRAALHRRGMDV-DPLCPVCANEEETVFHLFVSCPVAQNF 1011
A R++ LP ++ + +R +D+ D LCP C+ +EET HLF C Q+F
Sbjct: 479 FAGRLIRDRLPTKSNMRKRQIDINDSLCPFCSIKEETASHLFFDCSKTQHF 327
Score = 22.7 bits (47), Expect(2) = 1e-09
Identities = 6/17 (35%), Positives = 13/17 (76%)
Frame = -1
Query: 1046 WQAGVYALWEMRNRVVF 1062
W A +++W+ RN+++F
Sbjct: 204 WVALNWSIWQHRNKIIF 154
>BI702277
Length = 420
Score = 47.4 bits (111), Expect(2) = 1e-09
Identities = 22/54 (40%), Positives = 32/54 (58%)
Frame = +2
Query: 1136 GEVLASAAQYLLHAASAILGEALVFRWSMQLTIQMEFRRVLFETDCLQLFQLWK 1189
GEVLASA + S + E L F+WS+Q+ Q+ + V FETDC ++ W+
Sbjct: 2 GEVLASATRLEDQCYSPQIAEYLAFKWSIQMANQLLLQNVFFETDCREIASAWR 163
Score = 34.7 bits (78), Expect(2) = 1e-09
Identities = 20/60 (33%), Positives = 31/60 (51%)
Frame = +1
Query: 1192 PDGRSYLSSIVGDCFMLSRFFDYVDLSFVRRRGNSVADFLAGTASKYTDMVWLEEVPREV 1251
P +YL +I+ DC ++ D V L ++ GN VAD LA A D +E+ P ++
Sbjct: 172 PKDCTYLDAILQDCRDMTTHIDTVALVHTKKIGNIVADSLA*LAFVLVDQSGVEDAPPQL 351
>BE610670
Length = 368
Score = 61.2 bits (147), Expect = 3e-09
Identities = 39/122 (31%), Positives = 59/122 (47%), Gaps = 1/122 (0%)
Frame = -2
Query: 893 LKIHLPLNGSVDTLMWPETVDGNYCSKSGYIFVRKNFLGACSSTYSQPSLPAPLWKKFWR 952
++IHL S+D+L+W G Y +KS Y N L S ++ S K W
Sbjct: 337 VQIHL---SSLDSLIWRADPSGAYSTKSAY-----NLLKGEGSFDNEDSAS----KIIWN 194
Query: 953 TSAQPRCKEVAWRVVSSLLPIRAALHRRGMDVDPL-CPVCANEEETVFHLFVSCPVAQNF 1011
PR +WR+ + LP +A L RR +++ CP+C EEE V H+ SC ++
Sbjct: 193 LKIPPRTIAFSWRIFKNRLPTKANLRRRQVELPSYRCPLCDLEEENVGHIMFSCTRTRSL 14
Query: 1012 WF 1013
W+
Sbjct: 13 WW 8
>CO979823
Length = 853
Score = 60.8 bits (146), Expect = 4e-09
Identities = 47/172 (27%), Positives = 74/172 (42%), Gaps = 11/172 (6%)
Frame = -2
Query: 904 DTLMWPETVDGNYCSKSGYIFVRKNFLGACSSTYSQPSLPAPLWKKFWRTSAQPRCKEVA 963
D +W G+Y +KSGY + G + + + W+ + A
Sbjct: 702 DKWLWKPEPGGHYSTKSGYHVL----WGELTEEIQDAD-----FAEIWKLKIPTKAAVFA 550
Query: 964 WRVVSSLLPIRAALHRRGMDV-DPLCPVCANEEETVFHLFVSCPVAQNFWFGS------- 1015
WR+V LP ++ L RR + V D +CP+C N EE HLF +C W+ S
Sbjct: 549 WRLVRDRLPTKSNLRRRQVMVQDMVCPLCNNIEEGAAHLFFNCTKTLPLWWESMSWVNLK 370
Query: 1016 ---PLSLRVHGFSSMEDFLADFFRAADDDALALWQAGVYALWEMRNRVVFRD 1064
P + R H F +AD ++ W A + +W+ RN+VVF++
Sbjct: 369 TAMPQTPRQH-FLQYGTDIADGLKS--KRWKCWWIALTWTIWQHRNKVVFQN 223
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.324 0.139 0.438
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 64,253,017
Number of Sequences: 63676
Number of extensions: 1020606
Number of successful extensions: 6778
Number of sequences better than 10.0: 118
Number of HSP's better than 10.0 without gapping: 6605
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 6725
length of query: 1259
length of database: 12,639,632
effective HSP length: 108
effective length of query: 1151
effective length of database: 5,762,624
effective search space: 6632780224
effective search space used: 6632780224
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.5 bits)
S2: 65 (29.6 bits)
Lotus: description of TM0129.4


