

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0120.7
(373 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
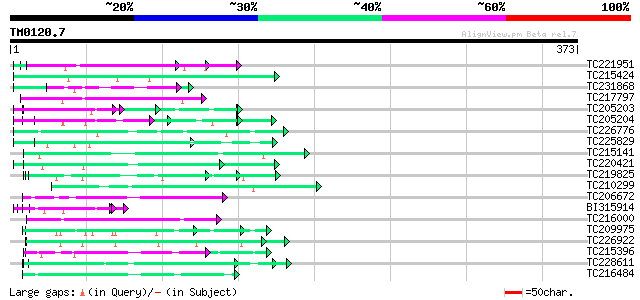
Score E
Sequences producing significant alignments: (bits) Value
TC221951 UP|NO75_SOYBN (P08297) Early nodulin 75 precursor (N-75... 74 8e-14
TC215424 similar to UP|Q9MA04 (Q9MA04) F20B17.16, partial (26%) 59 3e-09
TC231868 similar to UP|Q93424 (Q93424) C. elegans GRL-23 protein... 57 1e-08
TC217797 similar to UP|Q9LVI6 (Q9LVI6) Probable receptor-like pr... 56 2e-08
TC205203 similar to UP|Q39763 (Q39763) Proline-rich cell wall pr... 56 3e-08
TC205204 homologue to UP|Q43558 (Q43558) Proline rich protein pr... 55 5e-08
TC226776 similar to GB|AAP68287.1|31711862|BT008848 At1g02720 {A... 54 1e-07
TC225829 weakly similar to UP|Q9C5Q6 (Q9C5Q6) Fasciclin-like ara... 54 1e-07
TC215141 similar to GB|AAL32703.1|17065098|AY062625 nucleotide s... 53 2e-07
TC220421 weakly similar to UP|GP1_CHLRE (Q9FPQ6) Vegetative cell... 53 3e-07
TC219825 weakly similar to UP|MID2_YEAST (P36027) Mating process... 52 3e-07
TC210299 homologue to UP|EFT2_SOYBN (P46280) Elongation factor T... 52 4e-07
TC206672 weakly similar to UP|Q7UTL3 (Q7UTL3) 3-hydroxyisobutyra... 52 6e-07
BI315914 similar to PIR|T09854|T098 proline-rich cell wall prote... 52 6e-07
TC216000 similar to UP|O22253 (O22253) Photolyase/blue-light rec... 51 1e-06
TC209975 weakly similar to UP|Q7T9Y7 (Q7T9Y7) Odv-e66, partial (9%) 50 1e-06
TC226922 similar to UP|Q9LPQ3 (Q9LPQ3) F15H18.14, partial (54%) 50 2e-06
TC215396 weakly similar to UP|Q8H9F4 (Q8H9F4) 41 kD chloroplast ... 50 2e-06
TC228611 weakly similar to GB|CAA72487.1|1890317|ATP26A peroxida... 50 2e-06
TC216484 similar to UP|FLA1_ARATH (Q9FM65) Fasciclin-like arabin... 49 3e-06
>TC221951 UP|NO75_SOYBN (P08297) Early nodulin 75 precursor (N-75) (NGM-75),
complete
Length = 1179
Score = 74.3 bits (181), Expect = 8e-14
Identities = 43/152 (28%), Positives = 61/152 (39%), Gaps = 2/152 (1%)
Frame = +1
Query: 3 PLDQDHPASPSSEESAPASPDPSQQPPPQDPPSPAQSQPETSDDPPQQPPPQDPLQPEQS 62
P ++ P E P P + PP + P P + QP P QPP + P +
Sbjct: 241 PHEKPPPEYLPPHEKPPPEYQPPHEKPPHENP-PPEHQPPHEKPPEHQPPHEKPPPEYEP 417
Query: 63 LSESPDPSQQSTLQDPLLLEQALNETAGSSQQPPQESSQSERSPPQSRRPEHQATESRLP 122
E P P Q + P Q +E QPP E E PP + PEHQ + P
Sbjct: 418 PHEKPPPEYQPPHEKPPPEYQPPHEKPPPEYQPPHEKPPPEHQPPHEKPPEHQPPHEKPP 597
Query: 123 AQQRP--ESSQPEHQATESRSLEQQQPPSLTP 152
+ +P E PE+Q + + ++ PP P
Sbjct: 598 PEYQPPHEKPPPEYQPPQEKPPHEKPPPEYQP 693
Score = 65.1 bits (157), Expect = 5e-11
Identities = 41/150 (27%), Positives = 52/150 (34%)
Frame = +1
Query: 3 PLDQDHPASPSSEESAPASPDPSQQPPPQDPPSPAQSQPETSDDPPQQPPPQDPLQPEQS 62
P + P E P P + PP + P P P P QPP + P Q
Sbjct: 337 PPEHQPPHEKPPEHQPPHEKPPPEYEPPHEKPPPEYQPPHEKPPPEYQPPHEKPPPEYQP 516
Query: 63 LSESPDPSQQSTLQDPLLLEQALNETAGSSQQPPQESSQSERSPPQSRRPEHQATESRLP 122
E P P Q + P Q +E QPP E E PPQ + P + P
Sbjct: 517 PHEKPPPEHQPPHEKPPE-HQPPHEKPPPEYQPPHEKPPPEYQPPQEKPPHEKPPPEYQP 693
Query: 123 AQQRPESSQPEHQATESRSLEQQQPPSLTP 152
++P PEHQ + PP P
Sbjct: 694 PHEKP---PPEHQPPHEKPPPVYPPPYEKP 774
Score = 64.7 bits (156), Expect = 6e-11
Identities = 42/144 (29%), Positives = 59/144 (40%), Gaps = 3/144 (2%)
Frame = +1
Query: 12 PSSEESAPAS-PDPSQQPPPQDPPSPAQSQPETSDDPPQQPPPQDPLQPEQSLSESPDPS 70
P E++ P P P ++PPP+ P + PE PP + PP + PE P
Sbjct: 202 PPHEKTPPEYLPPPHEKPPPEYLPPHEKPPPEYQ--PPHEKPPHENPPPEHQPPHEKPPE 375
Query: 71 QQSTLQDPLLLEQALNETAGSSQQPPQESSQSERSPPQSR-RPEHQATESRLPAQ-QRPE 128
Q + P + +E QPP E E PP + PE+Q + P + Q P
Sbjct: 376 HQPPHEKPPPEYEPPHEKPPPEYQPPHEKPPPEYQPPHEKPPPEYQPPHEKPPPEHQPPH 555
Query: 129 SSQPEHQATESRSLEQQQPPSLTP 152
PEHQ + + QPP P
Sbjct: 556 EKPPEHQPPHEKPPPEYQPPHEKP 627
Score = 62.8 bits (151), Expect = 2e-10
Identities = 41/152 (26%), Positives = 57/152 (36%), Gaps = 2/152 (1%)
Frame = +1
Query: 3 PLDQDHPASPSSEESAPASPDPSQQPPPQDPPSPAQSQPETSDDPPQQPPPQDPLQPEQS 62
P+++ P P P P PPP+ P P + P PP + PP + L P
Sbjct: 109 PIEKPPTYEPPPFYKPPYYPPPVHHPPPEYQP-PHEKTPPEYLPPPHEKPPPEYLPPH-- 279
Query: 63 LSESPDPSQQSTLQDPLLLEQALNETAGSSQQPPQESSQSERSPPQSRRPEHQATESRLP 122
E P P Q ++PP E+ E PP + PEHQ + P
Sbjct: 280 --EKPPPEYQP-----------------PHEKPPHENPPPEHQPPHEKPPEHQPPHEKPP 402
Query: 123 AQQRP--ESSQPEHQATESRSLEQQQPPSLTP 152
+ P E PE+Q + + QPP P
Sbjct: 403 PEYEPPHEKPPPEYQPPHEKPPPEYQPPHEKP 498
Score = 48.5 bits (114), Expect = 5e-06
Identities = 32/130 (24%), Positives = 46/130 (34%), Gaps = 5/130 (3%)
Frame = +1
Query: 8 HPASPSSEESAPASPDPSQQPPPQDPPS---PAQSQPETSDDPPQQPPPQDPLQPEQSLS 64
H P + P P QPP + PP P + P P ++PPP+ E+
Sbjct: 487 HEKPPPEYQPPHEKPPPEHQPPHEKPPEHQPPHEKPPPEYQPPHEKPPPEYQPPQEKPPH 666
Query: 65 ESPDPSQQSTLQDPLLLEQALNETAGSSQQPPQESSQSERSPPQSRRPE--HQATESRLP 122
E P P Q + P Q +E PP E PP + P + + P
Sbjct: 667 EKPPPEYQPPHEKPPPEHQPPHEKPPPVYPPPYEKPPPVYEPPYEKPPPVVYPPPHEKPP 846
Query: 123 AQQRPESSQP 132
+ P +P
Sbjct: 847 IYEPPPLEKP 876
Score = 44.3 bits (103), Expect = 9e-05
Identities = 28/111 (25%), Positives = 39/111 (34%), Gaps = 1/111 (0%)
Frame = +1
Query: 3 PLDQDHPASPSSEESAPASPDPSQQPPPQDPPSPAQSQPETSDDPPQQPPPQDPLQPEQS 62
P ++ P E P P Q+ PP + P P P P QPP + P
Sbjct: 580 PHEKPPPEYQPPHEKPPPEYQPPQEKPPHEKPPPEYQPPHEKPPPEHQPPHEKPPPVYPP 759
Query: 63 LSESPDPSQQSTLQ-DPLLLEQALNETAGSSQQPPQESSQSERSPPQSRRP 112
E P P + + P ++ +E + PP E PP R P
Sbjct: 760 PYEKPPPVYEPPYEKPPPVVYPPPHEKPPIYEPPPLEKPPVYNPPPYGRYP 912
Score = 28.9 bits (63), Expect = 3.9
Identities = 18/66 (27%), Positives = 22/66 (33%), Gaps = 7/66 (10%)
Frame = +1
Query: 3 PLDQDHPASPSSEESAPA-------SPDPSQQPPPQDPPSPAQSQPETSDDPPQQPPPQD 55
P ++ P P E P P P PPP + P P P P PPP
Sbjct: 727 PHEKPPPVYPPPYEKPPPVYEPPYEKPPPVVYPPPHEKP-PIYEPPPLEKPPVYNPPPYG 903
Query: 56 PLQPEQ 61
P +
Sbjct: 904 RYPPSK 921
>TC215424 similar to UP|Q9MA04 (Q9MA04) F20B17.16, partial (26%)
Length = 1030
Score = 59.3 bits (142), Expect = 3e-09
Identities = 46/190 (24%), Positives = 75/190 (39%), Gaps = 15/190 (7%)
Frame = +3
Query: 3 PLDQDHPASPSSEESAPASPDPSQQPPPQDPPSPA--QSQPETSDDPPQQPPPQDPLQPE 60
P + HP P P P+ S QPPP PP+P S + S+ PP PPP P P
Sbjct: 105 PPNPHHPHYPYPPPPPPPPPEASYQPPPPPPPAPMYYPSNNQYSNQPPPPPPPLSPPPPP 284
Query: 61 QSLSESPDP-----SQQSTLQDPLLLEQALNETAG-------SSQQPPQESSQSERSPP- 107
+S P P +++ ++P + E + QQPP + PP
Sbjct: 285 PPVSPPPPPPVTQNNEERHFKEPSTSGRREYEHSNHGIAHKQHKQQPPVPVKKMNNGPPG 464
Query: 108 QSRRPEHQATESRLPAQQRPESSQPEHQATESRSLEQQQPPSLTPGRPASEFRLLRGSSS 167
++ E + + +++ + + Q ES++ Q+ L+ G+ R
Sbjct: 465 RAETDEEKRLRKKREFEKQRQEEKHRQQLKESQNTVLQKTHMLSSGKGHGMIAGSRMGER 644
Query: 168 TSVPSLESTR 177
S P L + R
Sbjct: 645 RSTPLLGAER 674
Score = 34.3 bits (77), Expect = 0.093
Identities = 33/140 (23%), Positives = 49/140 (34%), Gaps = 14/140 (10%)
Frame = +3
Query: 23 DPSQQPPPQDPPSPAQSQPETSDDP-----PQQP----PPQDPLQPEQSLSESPDPSQQS 73
DPS PQ PP+ Q + P P P PP P P ++ + P P +
Sbjct: 24 DPSTSSFPQIPPNSNYHQNQQHHVPYAPPNPHHPHYPYPPPPPPPPPEASYQPPPPPPPA 203
Query: 74 TLQDPLLLEQALNETAGSSQQPPQESSQSERSPPQSRRPEHQATESRLPAQQRPESSQPE 133
+ P + N+ + PP S PP S P T++ + S+
Sbjct: 204 PMYYP-----SNNQYSNQPPPPPPPLSPPPPPPPVSPPPPPPVTQNNEERHFKEPSTSGR 368
Query: 134 HQATES-----RSLEQQQPP 148
+ S +QQPP
Sbjct: 369 REYEHSNHGIAHKQHKQQPP 428
>TC231868 similar to UP|Q93424 (Q93424) C. elegans GRL-23 protein
(Corresponding sequence E02A10.2), partial (23%)
Length = 790
Score = 57.0 bits (136), Expect = 1e-08
Identities = 41/121 (33%), Positives = 46/121 (37%), Gaps = 2/121 (1%)
Frame = -3
Query: 3 PLDQDHPASPSSEESAPASPDPSQQPPPQDPPSPAQSQP--ETSDDPPQQPPPQDPLQPE 60
P P P P P QPPPQ PP PA P PP QPPPQ P P
Sbjct: 458 PQPPPQPPQPLLSNLFPHPPPQPPQPPPQPPPQPAPQPPLLIKFPQPPPQPPPQPPQPP- 282
Query: 61 QSLSESPDPSQQSTLQDPLLLEQALNETAGSSQQPPQESSQSERSPPQSRRPEHQATESR 120
L + P P Q Q PLL+ Q PPQ + Q + P P+ S
Sbjct: 281 -PLIKFPQPPPQPEPQPPLLII--------FPQPPPQPAPQPPQPPLSIAFPQPPMDSSD 129
Query: 121 L 121
L
Sbjct: 128 L 126
Score = 42.4 bits (98), Expect = 3e-04
Identities = 32/89 (35%), Positives = 39/89 (42%)
Frame = -3
Query: 25 SQQPPPQDPPSPAQSQPETSDDPPQQPPPQDPLQPEQSLSESPDPSQQSTLQDPLLLEQA 84
S + PPQ PP P QP S+ P PPPQ P P P P Q Q PLL++
Sbjct: 473 SIRSPPQPPPQP--PQPLLSNLFP-HPPPQPPQPP-------PQPPPQPAPQPPLLIK-- 330
Query: 85 LNETAGSSQQPPQESSQSERSPPQSRRPE 113
Q PPQ Q + PP + P+
Sbjct: 329 ------FPQPPPQPPPQPPQPPPLIKFPQ 261
>TC217797 similar to UP|Q9LVI6 (Q9LVI6) Probable receptor-like protein kinase
protein (AT3g17840/MEB5_6), partial (44%)
Length = 1612
Score = 56.2 bits (134), Expect = 2e-08
Identities = 43/131 (32%), Positives = 54/131 (40%), Gaps = 9/131 (6%)
Frame = -3
Query: 8 HPASPSSEESAPASPDPSQQPPPQDP--PSPAQSQPETSD-DPPQQPPPQDPLQP-EQSL 63
HP S AP S DPS PP P PS S P TS PP PPP P P +SL
Sbjct: 464 HPC*NSKP*EAPNSRDPSLPPPQSSP*IPSRKLSHPSTSSRPPP-APPPTPPCTPSRKSL 288
Query: 64 SESPDPSQQSTLQDPLLLEQALNETAGSSQQPPQESSQSERSPPQSRRP-----EHQATE 118
S P P + L + + S PP + PPQ R +H++
Sbjct: 287 SPKPPPKP*ANLPNQTPSPHSQKTPTSSRSLPPCYHHSQPQPPPQKRLQG*HWHDHRSRH 108
Query: 119 SRLPAQQRPES 129
R P +Q P++
Sbjct: 107 RRHPCRQEPQT 75
>TC205203 similar to UP|Q39763 (Q39763) Proline-rich cell wall protein,
partial (48%)
Length = 1076
Score = 55.8 bits (133), Expect = 3e-08
Identities = 44/146 (30%), Positives = 53/146 (36%), Gaps = 1/146 (0%)
Frame = +3
Query: 9 PASPSSEESAPASPDPSQQPPPQDPPSPAQ-SQPETSDDPPQQPPPQDPLQPEQSLSESP 67
P SS P+SP PSQ PPP P P+ S P S PP PPP P P SP
Sbjct: 318 PPVQSSPPPVPSSPPPSQSPPPASTPPPSPLSSPPPSSPPPSSPPPSSP-PPASPPPSSP 494
Query: 68 DPSQQSTLQDPLLLEQALNETAGSSQQPPQESSQSERSPPQSRRPEHQATESRLPAQQRP 127
P+ P ++ PP + PP P T + P P
Sbjct: 495 PPASPPPSSPP--------PSSPPPFSPPPATPPPATPPPAV--PPPSLTPTVTPLSSPP 644
Query: 128 ESSQPEHQATESRSLEQQQPPSLTPG 153
SS + S+ PSL PG
Sbjct: 645 VSSP---APSPSKVFAPALSPSLAPG 713
Score = 53.1 bits (126), Expect = 2e-07
Identities = 46/144 (31%), Positives = 50/144 (33%), Gaps = 1/144 (0%)
Frame = +3
Query: 10 ASPSSEESAPASPDPSQQPPP-QDPPSPAQSQPETSDDPPQQPPPQDPLQPEQSLSESPD 68
A ++PA P P PPP Q P P S P PP Q PP P LS SP
Sbjct: 261 AQAQGPSTSPAPPTPQASPPPVQSSPPPVPSSP-----PPSQSPPPASTPPPSPLS-SPP 422
Query: 69 PSQQSTLQDPLLLEQALNETAGSSQQPPQESSQSERSPPQSRRPEHQATESRLPAQQRPE 128
PS + S PP SS SPP S P S P P
Sbjct: 423 PS-----------------SPPPSSPPP--SSPPPASPPPSSPPPASPPPSSPPPSSPPP 545
Query: 129 SSQPEHQATESRSLEQQQPPSLTP 152
S P + PPSLTP
Sbjct: 546 FSPPPATPPPATPPPAVPPPSLTP 617
Score = 52.0 bits (123), Expect = 4e-07
Identities = 29/74 (39%), Positives = 33/74 (44%), Gaps = 1/74 (1%)
Frame = +3
Query: 3 PLDQDHPASPSSEESAPASPDPSQQPPPQDPP-SPAQSQPETSDDPPQQPPPQDPLQPEQ 61
PL P+SP P+SP P+ PP PP SP S P S PP PPP P P
Sbjct: 405 PLSSPPPSSPPPSSPPPSSPPPASPPPSSPPPASPPPSSPPPSSPPPFSPPPATP-PPAT 581
Query: 62 SLSESPDPSQQSTL 75
P PS T+
Sbjct: 582 PPPAVPPPSLTPTV 623
Score = 48.5 bits (114), Expect = 5e-06
Identities = 29/98 (29%), Positives = 36/98 (36%), Gaps = 1/98 (1%)
Frame = +3
Query: 3 PLDQDHPASPSSEESAPASPDPSQQPPPQDPPS-PAQSQPETSDDPPQQPPPQDPLQPEQ 61
P Q P + + S +SP PS PP PPS P + P S PP PPP P
Sbjct: 360 PPSQSPPPASTPPPSPLSSPPPSSPPPSSPPPSSPPPASPPPSSPPPASPPPSSPPPSSP 539
Query: 62 SLSESPDPSQQSTLQDPLLLEQALNETAGSSQQPPQES 99
P + P + +L T PP S
Sbjct: 540 PPFSPPPATPPPATPPPAVPPPSLTPTVTPLSSPPVSS 653
Score = 46.6 bits (109), Expect = 2e-05
Identities = 28/75 (37%), Positives = 32/75 (42%), Gaps = 6/75 (8%)
Frame = +3
Query: 3 PLDQDHPASPSSEESAPASPDPSQQPPPQDPP-SPAQSQPETSDDPPQQPP-----PQDP 56
P PASP PASP PS PP PP SP + P + PP PP P
Sbjct: 450 PPSSPPPASPPPSSPPPASPPPSSPPPSSPPPFSPPPATPPPATPPPAVPPPSLTPTVTP 629
Query: 57 LQPEQSLSESPDPSQ 71
L S +P PS+
Sbjct: 630 LSSPPVSSPAPSPSK 674
Score = 37.7 bits (86), Expect = 0.008
Identities = 23/70 (32%), Positives = 28/70 (39%), Gaps = 2/70 (2%)
Frame = +3
Query: 3 PLDQDHPASPSSEESAPASPDPSQQPPPQDPPSPAQSQPETSDDPPQQPPPQDP--LQPE 60
P P+SP PA+P P+ PP PPS + S P P P P
Sbjct: 510 PPSSPPPSSPPPFSPPPATPPPATPPPAVPPPSLTPTVTPLSSPPVSSPAPSPSKVFAPA 689
Query: 61 QSLSESPDPS 70
S S +P PS
Sbjct: 690 LSPSLAPGPS 719
>TC205204 homologue to UP|Q43558 (Q43558) Proline rich protein precursor,
partial (44%)
Length = 996
Score = 55.1 bits (131), Expect = 5e-08
Identities = 44/148 (29%), Positives = 54/148 (35%), Gaps = 3/148 (2%)
Frame = +1
Query: 9 PASPSSEESAPASPDPSQQPPPQDPPSPAQ-SQPETSDDPPQQPPPQDPLQPEQSLSESP 67
P SS P+SP P+Q PPP P PA S P + PP PPP P P SP
Sbjct: 226 PPIQSSPPPVPSSPPPAQSPPPASTPPPAPLSSPPPASPPPSSPPPASP-PPASPPPASP 402
Query: 68 DPSQQSTLQDPLLLEQALNETAGSSQQPPQESSQSERSPPQSRRPEHQATESRLPAQQRP 127
P+ P A P +S SPP + P + P P
Sbjct: 403 PPASPPPASPP---------PASPPPATPPPASPPPFSPPPATPPPATPPPALTPT---P 546
Query: 128 ESSQP--EHQATESRSLEQQQPPSLTPG 153
SS P ++ + PSL PG
Sbjct: 547 LSSPPATSPAPAPAKVVAPALSPSLAPG 630
Score = 52.0 bits (123), Expect = 4e-07
Identities = 32/94 (34%), Positives = 39/94 (41%), Gaps = 1/94 (1%)
Frame = +1
Query: 3 PLDQDHPASPSSEESAPASPDPSQQPPPQDPP-SPAQSQPETSDDPPQQPPPQDPLQPEQ 61
PL PASP PASP P+ PP PP SP + P + PP PPP P P
Sbjct: 313 PLSSPPPASPPPSSPPPASPPPASPPPASPPPASPPPASPPPASPPPATPPPASP-PPFS 489
Query: 62 SLSESPDPSQQSTLQDPLLLEQALNETAGSSQQP 95
+P P+ P L L+ +S P
Sbjct: 490 PPPATPPPA----TPPPALTPTPLSSPPATSPAP 579
Score = 50.8 bits (120), Expect = 1e-06
Identities = 41/137 (29%), Positives = 47/137 (33%), Gaps = 1/137 (0%)
Frame = +1
Query: 17 SAPASPDPSQQPPP-QDPPSPAQSQPETSDDPPQQPPPQDPLQPEQSLSESPDPSQQSTL 75
++PA P P PPP Q P P S P PP Q PP P LS P S +
Sbjct: 190 TSPAPPTPQSSPPPIQSSPPPVPSSP-----PPAQSPPPASTPPPAPLSSPPPASPPPSS 354
Query: 76 QDPLLLEQALNETAGSSQQPPQESSQSERSPPQSRRPEHQATESRLPAQQRPESSQPEHQ 135
P PP S PP S P S PA P +S P
Sbjct: 355 PPP--------------ASPPPASPPPASPPPASPPPASPPPASPPPATP-PPASPPPFS 489
Query: 136 ATESRSLEQQQPPSLTP 152
+ PP+LTP
Sbjct: 490 PPPATPPPATPPPALTP 540
Score = 48.5 bits (114), Expect = 5e-06
Identities = 48/173 (27%), Positives = 59/173 (33%), Gaps = 6/173 (3%)
Frame = +1
Query: 9 PASPSSEESAPASPDPSQQPPPQDP--PSPAQSQPETSDDPP---QQPPPQDPLQPEQSL 63
P++ + + +SP P Q PP P P PAQS P S PP PPP P P
Sbjct: 184 PSTSPAPPTPQSSPPPIQSSPPPVPSSPPPAQSPPPASTPPPAPLSSPPPASP-PPSSPP 360
Query: 64 SESPDPSQQSTLQDPLLLEQALNETAGSSQQPPQESSQSERSPPQSRRPEHQATESRLPA 123
SP P+ P PP S PP + P S PA
Sbjct: 361 PASPPPASPPPASPP-------------PASPPPASPPPASPPPATPPPASPPPFSPPPA 501
Query: 124 QQRPESSQPEHQATESRSLEQQQPPSLTPG-RPASEFRLLRGSSSTSVPSLES 175
P + P T S PP+ +P PA S PSL +
Sbjct: 502 TPPPATPPPALTPTPLSS-----PPATSPAPAPAKVVAPALSPSLAPGPSLST 645
Score = 45.8 bits (107), Expect = 3e-05
Identities = 31/107 (28%), Positives = 38/107 (34%), Gaps = 2/107 (1%)
Frame = +1
Query: 3 PLDQDHPASPSSEESAPASPDPSQQPPPQDPP--SPAQSQPETSDDPPQQPPPQDPLQPE 60
P P S+ AP S P PPP PP SP + P + PP PPP P P
Sbjct: 265 PPPAQSPPPASTPPPAPLSSPPPASPPPSSPPPASPPPASPPPASPPPASPPPASP-PPA 441
Query: 61 QSLSESPDPSQQSTLQDPLLLEQALNETAGSSQQPPQESSQSERSPP 107
+P P+ P T + PP + SPP
Sbjct: 442 SPPPATPPPASPPPFSPP-------PATPPPATPPPALTPTPLSSPP 561
>TC226776 similar to GB|AAP68287.1|31711862|BT008848 At1g02720 {Arabidopsis
thaliana;} , partial (68%)
Length = 1307
Score = 53.9 bits (128), Expect = 1e-07
Identities = 57/202 (28%), Positives = 78/202 (38%), Gaps = 21/202 (10%)
Frame = +2
Query: 3 PLDQDHPASPSSEESAPASPDPSQQPPPQDPPSPAQSQPETSDDPPQQPPPQDP--LQPE 60
PL P +P++ P SP PS PP PSP+ S + PP P P +
Sbjct: 608 PLVSAMPPTPTNAPPPP-SPPPSATPPSSTSPSPSTSSTSAAPSPPSTPSSNTPSARRTS 784
Query: 61 QSLSESPDPSQQSTLQDPLLLEQALNETAGSSQQPPQESSQSERSPPQSRRPEHQATESR 120
S S SP P+ + P +LN T+ S P+ S + SPP S +P + S
Sbjct: 785 SSTSSSPKPTSNPSSNPP-----SLN*TSRSITLIPR--SSAT*SPPPSDKPSN--NPST 937
Query: 121 LPAQQRPESSQPEHQATESRS------------------LEQQQPPS-LTPGRPASEFRL 161
P SS P + + + LE PPS TP P++
Sbjct: 938 TPEITSLTSSNPASRGLYTSTPIWFSSTTLPSSGAPV*VLEPLAPPSTATPTSPSTS--- 1108
Query: 162 LRGSSSTSVPSLESTRGGRAFA 183
+GS T S GGRA +
Sbjct: 1109PQGSGRTCASPARSRGGGRAIS 1174
Score = 40.0 bits (92), Expect = 0.002
Identities = 50/186 (26%), Positives = 68/186 (35%), Gaps = 27/186 (14%)
Frame = +2
Query: 14 SEESAPASPDPSQQPPPQDPPSPA------------QSQPETSDDPP----QQPPPQDPL 57
S S+P +PS P P PP+ + + P S PP PPP P
Sbjct: 491 SSFSSPLPFNPSTPPKPSAPPTTSTASSASLRLAFPSAPPLVSAMPPTPTNAPPPPSPPP 670
Query: 58 QPEQSLSESPDPSQQSTL------QDPLLLEQALNETAGSSQQPPQESSQSERSPPQSRR 111
S SP PS ST P + T+ S+ P+ +S +PP
Sbjct: 671 SATPPSSTSPSPSTSSTSAAPSPPSTPSSNTPSARRTSSSTSSSPKPTSNPSSNPPSLN* 850
Query: 112 PEHQATESRLPAQQRPESSQPEHQATESRSLEQQQPPSLTPGRPASEFRLLRGS-----S 166
T +P S P + + + S + SLT PAS R L S S
Sbjct: 851 TSRSIT--LIPRSSAT*SPPPSDKPSNNPS-TTPEITSLTSSNPAS--RGLYTSTPIWFS 1015
Query: 167 STSVPS 172
ST++PS
Sbjct: 1016STTLPS 1033
>TC225829 weakly similar to UP|Q9C5Q6 (Q9C5Q6) Fasciclin-like
arabinogalactan-protein 2, partial (57%)
Length = 1652
Score = 53.9 bits (128), Expect = 1e-07
Identities = 46/164 (28%), Positives = 62/164 (37%), Gaps = 4/164 (2%)
Frame = +2
Query: 17 SAPASPDPSQQPPPQDPPSPAQSQP----ETSDDPPQQPPPQDPLQPEQSLSESPDPSQQ 72
S+ S S PPP PPSP + P T PP P P++S + P PS
Sbjct: 17 SSSLSSSSSS*PPPPSPPSPPTTSPACWPHTPASPPSTTTSPSPTWPKRSTAARPSPSWP 196
Query: 73 STLQDPLLLEQALNETAGSSQQPPQESSQSERSPPQSRRPEHQATESRLPAQQRPESSQP 132
ST+ + + + S P SS + +P S R S P RP + P
Sbjct: 197 STMPPCPPSSTSTSPSPPSKTSSPSTSSSTTSAPKSSTRSTTAPPSS--PPCSRPPAPPP 370
Query: 133 EHQATESRSLEQQQPPSLTPGRPASEFRLLRGSSSTSVPSLEST 176
AT + P+ P R AS +T+ PS ST
Sbjct: 371 APPATST-------SPTSRPARSASP------PKTTTAPSTPST 463
Score = 43.5 bits (101), Expect = 2e-04
Identities = 34/130 (26%), Positives = 47/130 (36%), Gaps = 10/130 (7%)
Frame = +2
Query: 3 PLDQDHPASPSSEESAPASPD-----PSQQPPPQDPPSPAQSQPETSDDPPQQP-----P 52
P P++ SS SAP S P PP PP+P + P TS P +P P
Sbjct: 248 PSKTSSPSTSSSTTSAPKSSTRSTTAPPSSPPCSRPPAPPPAPPATSTSPTSRPARSASP 427
Query: 53 PQDPLQPEQSLSESPDPSQQSTLQDPLLLEQALNETAGSSQQPPQESSQSERSPPQSRRP 112
P+ P + +P +T T PP S+ S P ++ RP
Sbjct: 428 PKTTTAPSTPSTSNPSRKCPTTSPSSKSAPPLAPPTPRHPPPPPPPST*SPLCPNKAARP 607
Query: 113 EHQATESRLP 122
E+ P
Sbjct: 608 SRTF*EAPRP 637
Score = 37.4 bits (85), Expect = 0.011
Identities = 24/104 (23%), Positives = 38/104 (36%), Gaps = 5/104 (4%)
Frame = +2
Query: 9 PASPSSEESAPASPDPSQQPPPQD-----PPSPAQSQPETSDDPPQQPPPQDPLQPEQSL 63
P PSS ++P+ P + P P S +S PP PP P P +
Sbjct: 209 PCPPSSTSTSPSPPSKTSSPSTSSSTTSAPKSSTRSTTAPPSSPPCSRPPAPPPAPPATS 388
Query: 64 SESPDPSQQSTLQDPLLLEQALNETAGSSQQPPQESSQSERSPP 107
+ +S + T+ S++ P S S+ +PP
Sbjct: 389 TSPTSRPARSASPPKTTTAPSTPSTSNPSRKCPTTSPSSKSAPP 520
>TC215141 similar to GB|AAL32703.1|17065098|AY062625 nucleotide sugar
epimerase-like protein {Arabidopsis thaliana;} , partial
(89%)
Length = 1743
Score = 53.1 bits (126), Expect = 2e-07
Identities = 56/203 (27%), Positives = 77/203 (37%), Gaps = 15/203 (7%)
Frame = +2
Query: 10 ASPSSEESA----PASPDPSQQPPPQDPP-SPAQSQPETSDDPPQQPPPQDPLQPEQSLS 64
+SPS S PAS P + P P PP S A S +PP PP L+P S +
Sbjct: 527 SSPSPTSSTWPHRPASATPCRTPNPTSPPTSLASSTFSRLPNPPTPNPPLSGLRPAPSTA 706
Query: 65 ESPDPSQQSTLQDPLLLEQALNETAGSSQQPPQESSQSERSPPQSRRPEHQATESRLPAQ 124
+P + L+ ++ + PP + R +S P T S P
Sbjct: 707 STP---------------KTLSPSSTAPTSPPASTPPPRRPARKSPTPTTTFTASPSPDS 841
Query: 125 QRPESSQPEHQATESRSLEQQQPPSLTPGRPASEFRLLRGS----------SSTSVPSLE 174
S+ P T L P + + GRP++ R RGS +S+ V S
Sbjct: 842 VSSPSTAPGEDPT---WLTSSSPKTSSMGRPSTSTRRRRGSRWRVTLPTSTTSSRVASAP 1012
Query: 175 STRGGRAFARLQESQPVPSLEST 197
ST RA S +PS E T
Sbjct: 1013STPPRRAPGAAARSGALPSSECT 1081
Score = 36.2 bits (82), Expect = 0.025
Identities = 23/79 (29%), Positives = 31/79 (39%)
Frame = +2
Query: 3 PLDQDHPASPSSEESAPASPDPSQQPPPQDPPSPAQSQPETSDDPPQQPPPQDPLQPEQS 62
P P S S S+ +SP + PP PP+ + T+ PP P P +S
Sbjct: 110 PNSSSAPLSWSHSSSSSSSPSTTLPSPPPTPPTATSTPTPTTSSPP------PPSAPGRS 271
Query: 63 LSESPDPSQQSTLQDPLLL 81
S +P P T L L
Sbjct: 272 RSATPPPPAAPTASPSLSL 328
Score = 30.4 bits (67), Expect = 1.3
Identities = 19/62 (30%), Positives = 26/62 (41%), Gaps = 4/62 (6%)
Frame = +2
Query: 13 SSEESAPAS----PDPSQQPPPQDPPSPAQSQPETSDDPPQQPPPQDPLQPEQSLSESPD 68
S + +AP+S P+ S P S + S P T+ P PP P + S P
Sbjct: 71 SGDSTAPSSSTPPPNSSSAPLSWSHSSSSSSSPSTTLPSPPPTPPTATSTPTPTTSSPPP 250
Query: 69 PS 70
PS
Sbjct: 251 PS 256
>TC220421 weakly similar to UP|GP1_CHLRE (Q9FPQ6) Vegetative cell wall
protein gp1 precursor (Hydroxyproline-rich glycoprotein
1), partial (13%)
Length = 1409
Score = 52.8 bits (125), Expect = 3e-07
Identities = 46/180 (25%), Positives = 68/180 (37%), Gaps = 5/180 (2%)
Frame = +3
Query: 3 PLDQDHPASPSSEESAPASPDPSQQPPPQDPPSPAQSQPETSDDP---PQQPPPQDPLQP 59
PL + P + ++P P+ PP PPS P + P P PP P
Sbjct: 168 PLWTQNKQQPLNPSTSPPHRTPAPSPPSTMPPSATPPSPSATSSPSASPTAPPTSPSPSP 347
Query: 60 EQSLSESPDPSQQSTLQDPLLLEQALNETAGSSQQPPQESSQSERSPPQSRRPEHQATES 119
S S PS ST P + + S PP +S + + P S P T
Sbjct: 348 PSSPSPPSAPSPSSTAPSP----PSASPPTSPSPSPPSPASTASANSPASGSP----TSR 503
Query: 120 RLPAQQRPES-SQPEHQATESRSLEQQQP-PSLTPGRPASEFRLLRGSSSTSVPSLESTR 177
P P S S+P ++ S + +P PS TP P +S TS+ + +++
Sbjct: 504 TSPTSPSPTSTSKPPAPSSSSPA*PSSKPSPSPTPISPDPSPATSTPTSHTSISPITASK 683
Score = 47.4 bits (111), Expect = 1e-05
Identities = 37/137 (27%), Positives = 53/137 (38%), Gaps = 5/137 (3%)
Frame = +3
Query: 10 ASPSSEESAPA---SPDPSQQPPPQDPPSPAQSQPET-SDDPPQQPPPQDPLQPEQSLSE 65
+SPS+ +AP SP P P P PSP+ + P S PP P P P + +
Sbjct: 294 SSPSASPTAPPTSPSPSPPSSPSPPSAPSPSSTAPSPPSASPPTSPSPSPPSPASTASAN 473
Query: 66 SPDPSQQSTLQDPLLLEQALNETAGSSQQPPQESSQSERSPPQSRRPE-HQATESRLPAQ 124
SP ++ P + + S+ +PP SS S P P + PA
Sbjct: 474 SPASGSPTSRTSP------TSPSPTSTSKPPAPSSSSPA*PSSKPSPSPTPISPDPSPAT 635
Query: 125 QRPESSQPEHQATESRS 141
P S T S++
Sbjct: 636 STPTSHTSISPITASKA 686
>TC219825 weakly similar to UP|MID2_YEAST (P36027) Mating process protein
MID2 (Serine-rich protein SMS1) (Protein kinase A
interference protein), partial (19%)
Length = 862
Score = 52.4 bits (124), Expect = 3e-07
Identities = 50/168 (29%), Positives = 63/168 (36%), Gaps = 2/168 (1%)
Frame = +2
Query: 13 SSEESAPASPDPSQQPPPQDPPSPAQSQPETSDDPPQQPPPQDPLQPEQSLSESPDPSQQ 72
SS S+ +SP PS PPP PSP P T P PPP P S S SP
Sbjct: 38 SSSTSSSSSPSPSSPPPPPSSPSP----PSTPPRPSPSPPPSPPSPASSSASSSPSYGSP 205
Query: 73 STLQDPLLLEQALNETAGSSQQPPQESSQSERSPPQSRRPEHQATESRLPAQQRPESSQP 132
S + + T SS P SS S S P + + S + P S+ P
Sbjct: 206 S----------S*SSTTPSSSSP--WSS*SSPSTPTTPSSSSSLSSSSSLSF*SPTSTSP 349
Query: 133 EHQATESRSLEQQQPPSLTP--GRPASEFRLLRGSSSTSVPSLESTRG 178
T S P + +P P + R GS +S P + S G
Sbjct: 350 P-SGTSPPSSPSSSPSTASPP*RSPTTSSRAGSGSPLSSSPPIWSPAG 490
Score = 42.7 bits (99), Expect = 3e-04
Identities = 40/138 (28%), Positives = 49/138 (34%), Gaps = 16/138 (11%)
Frame = +2
Query: 11 SPSSEESAPASPDPSQQPP------PQDPPSPAQSQPETSDD-------PPQQPPPQDPL 57
SPSS P+SP P PP P PPSPA S +S P P
Sbjct: 68 SPSSPPPPPSSPSPPSTPPRPSPSPPPSPPSPASSSASSSPSYGSPSS*SSTTPSSSSPW 247
Query: 58 QPEQSLSESPDPSQQSTLQDPLLLEQALNETAGSSQQPPQES---SQSERSPPQSRRPEH 114
S S PS S+L +L+ + +S PP + S SP + P
Sbjct: 248 SS*SSPSTPTTPSSSSSLSS----SSSLSF*SPTSTSPPSGTSPPSSPSSSPSTASPP*R 415
Query: 115 QATESRLPAQQRPESSQP 132
T S P SS P
Sbjct: 416 SPTTSSRAGSGSPLSSSP 469
Score = 41.2 bits (95), Expect = 8e-04
Identities = 36/143 (25%), Positives = 44/143 (30%)
Frame = +2
Query: 10 ASPSSEESAPASPDPSQQPPPQDPPSPAQSQPETSDDPPQQPPPQDPLQPEQSLSESPDP 69
+S SS + P PS PP PP P+ S P + P P S S P
Sbjct: 50 SSSSSPSPSSPPPPPSSPSPPSTPPRPSPSPPPSPPSPASSSASSSPSYGSPSS*SSTTP 229
Query: 70 SQQSTLQDPLLLEQALNETAGSSQQPPQESSQSERSPPQSRRPEHQATESRLPAQQRPES 129
S S T SS SS P S P + P+ +
Sbjct: 230 SSSSPWSS---*SSPSTPTTPSSSSSLSSSSSLSF*SPTSTSPPSGTSPPSSPSSSPSTA 400
Query: 130 SQPEHQATESRSLEQQQPPSLTP 152
S P T S P S +P
Sbjct: 401 SPP*RSPTTSSRAGSGSPLSSSP 469
>TC210299 homologue to UP|EFT2_SOYBN (P46280) Elongation factor Tu,
chloroplast precursor (EF-Tu), complete
Length = 1615
Score = 52.0 bits (123), Expect = 4e-07
Identities = 45/186 (24%), Positives = 68/186 (36%), Gaps = 8/186 (4%)
Frame = +2
Query: 28 PPPQDPPSPAQSQPETSDDPPQQPPPQDPLQPEQSLSESPDPSQQSTLQDPLLLEQALNE 87
PPP P P+ + P++S P PPP P S P+ S+ + P +
Sbjct: 110 PPPHTSPLPSSTPPQSSTSPLPTPPPAAAPSP------SAPPAASSSARSP--TSTSAPS 265
Query: 88 TAGSSQQPPQESSQSERSPPQSRRPEHQATESRLPAQQRPESSQPEHQATESRSLEQQQP 147
++ +PP SPP + P T S P + P +S + +R P
Sbjct: 266 ATSTTVRPPSPPPSPWPSPPSAIVPLKNTTRSTPPPRSAPAASPSTPPPSNTRPRTATTP 445
Query: 148 PSLTPGRPASEF-------RLLRGSSSTSVPSLESTRGGRAFARLQESQ-PVPSLESTRG 199
S P P + R SSS+ P+ + + F+ +S P S ST
Sbjct: 446 TSTAPATPTTSKT*SPAPPRWTAPSSSSPAPTAQCPKPKNTFSWPNKSAFPTLSSSSTNR 625
Query: 200 GRAFAR 205
R R
Sbjct: 626 TRLMTR 643
Score = 28.1 bits (61), Expect = 6.7
Identities = 38/164 (23%), Positives = 54/164 (32%), Gaps = 10/164 (6%)
Frame = +2
Query: 65 ESPDPSQQSTLQDPLLLEQALNETAGSSQQPPQESSQSERSPPQSRR-------PEHQAT 117
+ P S + LL + + + PP S +PPQS P +
Sbjct: 23 QQPQSSHTLLMSTSLLHPPPIISSLKPTNPPPHTSPLPSSTPPQSSTSPLPTPPPAAAPS 202
Query: 118 ESRLPA---QQRPESSQPEHQATESRSLEQQQPPSLTPGRPASEFRLLRGSSSTSVPSLE 174
S PA R +S AT + PPS P P++ L + ST P
Sbjct: 203 PSAPPAASSSARSPTSTSAPSATSTTVRPPSPPPSPWPSPPSAIVPLKNTTRSTPPP--- 373
Query: 175 STRGGRAFARLQESQPVPSLESTRGGRAFARLQESQPVPSLEST 218
R+ S P PS +TR A + P+ T
Sbjct: 374 -----RSAPAASPSTPPPS--NTRPRTATTPTSTAPATPTTSKT 484
>TC206672 weakly similar to UP|Q7UTL3 (Q7UTL3) 3-hydroxyisobutyrate
dehydrogenase , partial (76%)
Length = 1230
Score = 51.6 bits (122), Expect = 6e-07
Identities = 38/135 (28%), Positives = 57/135 (42%)
Frame = +2
Query: 9 PASPSSEESAPASPDPSQQPPPQDPPSPAQSQPETSDDPPQQPPPQDPLQPEQSLSESPD 68
P +P+S S PA+P PS P PP P+ S S P PPP P QP + S
Sbjct: 125 PCAPTS--SVPATPSPSSTAP---PPRPSPS----STSGPTSPPPPTPSQPAPTSSSPSS 277
Query: 69 PSQQSTLQDPLLLEQALNETAGSSQQPPQESSQSERSPPQSRRPEHQATESRLPAQQRPE 128
+ +++ Q + T + PP + S + P P + + LP + +
Sbjct: 278 ATPRTSAQS--------SSTPPPAPSPPSAPAASSST*PPPNPPSPSKSRTPLPPKAATQ 433
Query: 129 SSQPEHQATESRSLE 143
S+ P AT +R E
Sbjct: 434 STPPCPAATAARRTE 478
Score = 32.7 bits (73), Expect = 0.27
Identities = 20/70 (28%), Positives = 30/70 (42%), Gaps = 7/70 (10%)
Frame = +2
Query: 11 SPSSEESAPASPDPSQQP-------PPQDPPSPAQSQPETSDDPPQQPPPQDPLQPEQSL 63
S S + P +P P P PP +PPSP++S+ PP+ P P +
Sbjct: 293 SAQSSSTPPPAPSPPSAPAASSST*PPPNPPSPSKSRTPL---PPKAATQSTPPCPAATA 463
Query: 64 SESPDPSQQS 73
+ +P Q S
Sbjct: 464 ARRTEP*QSS 493
Score = 30.8 bits (68), Expect = 1.0
Identities = 30/112 (26%), Positives = 36/112 (31%)
Frame = +2
Query: 190 PVPSLESTRGGRAFARLQESQPVPSLESTRGGRAFARLQESQPVPSLESTRGGRAASASR 249
P PS ST G + SQP P+ S Q S P S AAS+S
Sbjct: 185 PRPSPSSTSGPTSPPPPTPSQPAPTSSSPSSATPRTSAQSSSTPPPAPSPPSAPAASSST 364
Query: 250 QPQQEQSLEQGRRQPQHERQPEQGRRQPQQERQLEQGRRQPQQEQQLGRERR 301
P R P + Q P R +P Q G+ RR
Sbjct: 365 *PPPNPPSPSKSRTPLPPKAATQS--TPPCPAATAARRTEP*QSSPAGKNRR 514
Score = 30.4 bits (67), Expect = 1.3
Identities = 27/81 (33%), Positives = 31/81 (37%)
Frame = +2
Query: 91 SSQQPPQESSQSERSPPQSRRPEHQATESRLPAQQRPESSQPEHQATESRSLEQQQPPSL 150
SS PP S S S P S P P+Q P SS P AT S + P
Sbjct: 167 SSTAPPPRPSPSSTSGPTSPPPP-------TPSQPAPTSSSPS-SATPRTSAQSSSTPPP 322
Query: 151 TPGRPASEFRLLRGSSSTSVP 171
P P++ SSST P
Sbjct: 323 APSPPSAP----AASSST*PP 373
>BI315914 similar to PIR|T09854|T098 proline-rich cell wall protein - upland
cotton, partial (37%)
Length = 246
Score = 51.6 bits (122), Expect = 6e-07
Identities = 27/69 (39%), Positives = 32/69 (46%), Gaps = 1/69 (1%)
Frame = +1
Query: 3 PLDQDHPASPSSEESAPASPDPSQQPPPQDPP-SPAQSQPETSDDPPQQPPPQDPLQPEQ 61
PL P+SP P+SP P+ PP PP SP S P S PP PPP P P
Sbjct: 40 PLSSPPPSSPPPSSPPPSSPPPASPPPSSPPPASPPPSSPPPSTPPPSSPPPFSP-PPAT 216
Query: 62 SLSESPDPS 70
+P P+
Sbjct: 217 PPPATPPPA 243
Score = 47.0 bits (110), Expect = 1e-05
Identities = 27/63 (42%), Positives = 30/63 (46%), Gaps = 1/63 (1%)
Frame = +1
Query: 9 PASPSSEESAPASPDPSQQPPPQDPP-SPAQSQPETSDDPPQQPPPQDPLQPEQSLSESP 67
P SP S P+SP PS PP PP SP S P + PP PPP P P SP
Sbjct: 31 PPSPLSSPP-PSSPPPSSPPPSSPPPASPPPSSPPPASPPPSSPPPSTP-PPSSPPPFSP 204
Query: 68 DPS 70
P+
Sbjct: 205PPA 213
Score = 46.2 bits (108), Expect = 2e-05
Identities = 26/70 (37%), Positives = 29/70 (41%), Gaps = 5/70 (7%)
Frame = +1
Query: 14 SEESAPAS---PDPSQQPPPQDPP--SPAQSQPETSDDPPQQPPPQDPLQPEQSLSESPD 68
S+ PAS P P PPP PP SP S P + PP PPP P P +P
Sbjct: 1 SQSPPPASTPPPSPLSSPPPSSPPPSSPPPSSPPPASPPPSSPPPASP-PPSSPPPSTPP 177
Query: 69 PSQQSTLQDP 78
PS P
Sbjct: 178PSSPPPFSPP 207
Score = 45.4 bits (106), Expect = 4e-05
Identities = 25/65 (38%), Positives = 29/65 (44%), Gaps = 1/65 (1%)
Frame = +1
Query: 6 QDHPASPSSEESAPASPDPSQQPPPQDPP-SPAQSQPETSDDPPQQPPPQDPLQPEQSLS 64
Q P + + S +SP PS PP PP SP + P S PP PPP P P
Sbjct: 4 QSPPPASTPPPSPLSSPPPSSPPPSSPPPSSPPPASPPPSSPPPASPPPSSP-PPSTPPP 180
Query: 65 ESPDP 69
SP P
Sbjct: 181SSPPP 195
>TC216000 similar to UP|O22253 (O22253) Photolyase/blue-light receptor
(Photolyase/blue light photoreceptor PHR2), partial
(84%)
Length = 1641
Score = 50.8 bits (120), Expect = 1e-06
Identities = 34/128 (26%), Positives = 52/128 (40%)
Frame = +3
Query: 12 PSSEESAPASPDPSQQPPPQDPPSPAQSQPETSDDPPQQPPPQDPLQPEQSLSESPDPSQ 71
P ++ S+ + P P PP P P+ +PP P P PL P + + P+
Sbjct: 69 PFNQPSSKSQPKPHPSHTFLSPP-PLLLLPKPPSNPPSPPTPSTPLSPSLPTAPATLPTP 245
Query: 72 QSTLQDPLLLEQALNETAGSSQQPPQESSQSERSPPQSRRPEHQATESRLPAQQRPESSQ 131
+ P + ++ S PP ++ S SP + P T SR PA RP +
Sbjct: 246 PPSAAPPSSGSATTSASSTMSASPPPTTTPSPSSPSTASTP--PTTASRHPASTRPAPTA 419
Query: 132 PEHQATES 139
P +T S
Sbjct: 420 PPSSSTPS 443
Score = 39.7 bits (91), Expect = 0.002
Identities = 54/210 (25%), Positives = 72/210 (33%), Gaps = 10/210 (4%)
Frame = +3
Query: 20 ASPDPSQQPPPQDPPSPAQSQPETSD---DPPQ-----QPPPQDPLQPEQSLSESPD-PS 70
++P S QPP P S +Q +P S PP +PP P P S SP P+
Sbjct: 42 SNPRISHQPPFNQPSSKSQPKPHPSHTFLSPPPLLLLPKPPSNPPSPPTPSTPLSPSLPT 221
Query: 71 QQSTLQDPLLLEQALNETAGSSQQPPQESSQSERSPPQSRRPEHQATESRLPAQQRPESS 130
+TL P S+ P S+ + S S P P P S
Sbjct: 222 APATLPTP----------PPSAAPPSSGSATTSASSTMSASP---------PPTTTPSPS 344
Query: 131 QPEHQATESRSLEQQQPPSLTPGRPASEFRLLRGSSSTSVPSLESTRGGRAFARLQESQP 190
P +T PP+ PAS S+S PS S R A P
Sbjct: 345 SPSTAST---------PPTTASRHPASTRPAPTAPPSSSTPSPTSAAASRRAA------P 479
Query: 191 VPSLESTRGGRA-FARLQESQPVPSLESTR 219
+ S S R ++ + S P P + R
Sbjct: 480 ISSCASGSPRRCWWSSPRPSVPTPCTPTAR 569
Score = 38.5 bits (88), Expect = 0.005
Identities = 30/110 (27%), Positives = 41/110 (37%), Gaps = 3/110 (2%)
Frame = +3
Query: 3 PLDQDHPASPSS--EESAPASPDPSQQPPPQD-PPSPAQSQPETSDDPPQQPPPQDPLQP 59
P + P +PS+ S P +P PPP PPS + S PPP P
Sbjct: 162 PSNPPSPPTPSTPLSPSLPTAPATLPTPPPSAAPPSSGSATTSASSTMSASPPPTTTPSP 341
Query: 60 EQSLSESPDPSQQSTLQDPLLLEQALNETAGSSQQPPQESSQSERSPPQS 109
+ S P+ S + P A SS P ++ S R+ P S
Sbjct: 342 SSPSTASTPPTTAS--RHPASTRPAPTAPPSSSTPSPTSAAASRRAAPIS 485
Score = 31.6 bits (70), Expect = 0.60
Identities = 14/25 (56%), Positives = 16/25 (64%)
Frame = -2
Query: 17 SAPASPDPSQQPPPQDPPSPAQSQP 41
S P SP PS PPP PPSP + +P
Sbjct: 638 SPPHSP-PSSPPPPSPPPSPRRERP 567
>TC209975 weakly similar to UP|Q7T9Y7 (Q7T9Y7) Odv-e66, partial (9%)
Length = 537
Score = 50.4 bits (119), Expect = 1e-06
Identities = 48/181 (26%), Positives = 72/181 (39%), Gaps = 17/181 (9%)
Frame = +1
Query: 9 PASPSSEESAPASPDPSQQPP----PQDPPSPAQSQPETSDDPPQQPPP--QDPLQPEQS 62
P +PS + P +P P PP P PSP+ S P PP+ P P Q P P
Sbjct: 4 PKTPSPIYAPPIAPSPGNHPPYVPTPPKTPSPSYSPPNV-PTPPKTPSPSSQPPFTPTPP 180
Query: 63 LSESPD------PSQQSTLQDPLLLEQALNETAGSSQQP-----PQESSQSERSPPQSRR 111
+ SP P+ S + P + N + +SQ P P +S S PP
Sbjct: 181 KTPSPTSQPPYIPTPPSPISQPPSIATPPNTLSPTSQPPYPSTIPPSTSLSPSYPPS--- 351
Query: 112 PEHQATESRLPAQQRPESSQPEHQATESRSLEQQQPPSLTPGRPASEFRLLRGSSSTSVP 171
P + + P+ P +S P + S + P + TP +S G+S+ + P
Sbjct: 352 PSPSSAPTYPPSYLAPATSPP--SPSTSATTPTISPAATTPSPSSSS---SSGASNETTP 516
Query: 172 S 172
+
Sbjct: 517 A 519
Score = 45.4 bits (106), Expect = 4e-05
Identities = 39/149 (26%), Positives = 50/149 (33%), Gaps = 4/149 (2%)
Frame = +1
Query: 11 SPSSEESAPASPDPSQQPP----PQDPPSPAQSQPETSDDPPQQPPPQDPLQPEQSLSES 66
SP + + P +P PS QPP P PSP P P P P +LS +
Sbjct: 106 SPPNVPTPPKTPSPSSQPPFTPTPPKTPSPTSQPPYIPTPPSPISQPPSIATPPNTLSPT 285
Query: 67 PDPSQQSTLQDPLLLEQALNETAGSSQQPPQESSQSERSPPQSRRPEHQATESRLPAQQR 126
P ST+ T+ S PP S S + P S + S +
Sbjct: 286 SQPPYPSTIPP---------STSLSPSYPPSPSPSSAPTYPPSYLAPATSPPSPSTSATT 438
Query: 127 PESSQPEHQATESRSLEQQQPPSLTPGRP 155
P S + S S TP RP
Sbjct: 439 PTISPAATTPSPSSSSSSGASNETTPARP 525
Score = 44.3 bits (103), Expect = 9e-05
Identities = 39/132 (29%), Positives = 51/132 (38%), Gaps = 16/132 (12%)
Frame = +1
Query: 9 PASPSSEESAPASPDPSQQPPPQD-------PPSPAQSQPETSDDPP------QQPPPQD 55
P +PS P +P P + P P PPSP SQP + PP QPP
Sbjct: 130 PKTPSPSSQPPFTPTPPKTPSPTSQPPYIPTPPSPI-SQPPSIATPPNTLSPTSQPPYPS 306
Query: 56 PLQPEQSLSES--PDPSQQST-LQDPLLLEQALNETAGSSQQPPQESSQSERSPPQSRRP 112
+ P SLS S P PS S P L A + + S+ S + +P S
Sbjct: 307 TIPPSTSLSPSYPPSPSPSSAPTYPPSYLAPATSPPSPSTSATTPTISPAATTPSPSSSS 486
Query: 113 EHQATESRLPAQ 124
A+ PA+
Sbjct: 487 SSGASNETTPAR 522
>TC226922 similar to UP|Q9LPQ3 (Q9LPQ3) F15H18.14, partial (54%)
Length = 1190
Score = 50.1 bits (118), Expect = 2e-06
Identities = 51/185 (27%), Positives = 69/185 (36%), Gaps = 12/185 (6%)
Frame = +2
Query: 12 PSSEESA-PASPDPSQQPPPQDPPSPAQSQPETSDDPPQQPPPQDPLQPEQSLSESPDPS 70
PSS +A P S PS+ PP SP+ S P PP PP PL P S S
Sbjct: 77 PSSATAATPTSVSPSRSPPSAASASPSPSLPP----PPPNQPPATPLPPPTSRS------ 226
Query: 71 QQSTLQDPLLLEQALNETAGSSQQPPQESSQSERSPPQSRRPEHQATESRLPAQQRPESS 130
P L + + S+ +PP ++S S P P A P+++ P S+
Sbjct: 227 ------SPSLATETAAPSTKSATRPPPPPTRSRSSTPTPTPPGAAA-----PSRKPPSSA 373
Query: 131 QPEHQATESRSLEQQQPPSLT-----------PGRPASEFRLLRGSSSTSVPSLESTRGG 179
T S S + P +T P RP S S + + S+RG
Sbjct: 374 ALPTAPTLSASTAPSKSPPVTWRSSWSTWTAAPSRPPSPPAARSRKSDSPRWRVTSSRGS 553
Query: 180 RAFAR 184
R R
Sbjct: 554 RTSTR 568
Score = 45.1 bits (105), Expect = 5e-05
Identities = 49/181 (27%), Positives = 71/181 (39%), Gaps = 22/181 (12%)
Frame = +2
Query: 11 SPSSEESAPASPDPSQQPPPQD-------PPSPAQSQPETSDD---PPQQPPPQDPLQPE 60
S S +A ASP PS PPP + PP ++S P + + P + + P P
Sbjct: 119 SRSPPSAASASPSPSLPPPPPNQPPATPLPPPTSRSSPSLATETAAPSTKSATRPPPPPT 298
Query: 61 QSLSESPD--------PSQQSTLQDPLLLEQALNETAGSSQQPP--QESSQSERSPPQSR 110
+S S +P PS++ L L+ + S+ PP SS S + SR
Sbjct: 299 RSRSSTPTPTPPGAAAPSRKPPSSAALPTAPTLSASTAPSKSPPVTWRSSWSTWTAAPSR 478
Query: 111 RPEHQATESRL--PAQQRPESSQPEHQATESRSLEQQQPPSLTPGRPASEFRLLRGSSST 168
P A SR + R SS+ +T + SL P + P RL S+
Sbjct: 479 PPSPPAARSRKSDSPRWRVTSSRGSRTSTRATSLTAT*SPRTS**TPKGMLRLQTSESAN 658
Query: 169 S 169
S
Sbjct: 659 S 661
>TC215396 weakly similar to UP|Q8H9F4 (Q8H9F4) 41 kD chloroplast nucleoid DNA
binding protein (CND41), partial (69%)
Length = 1808
Score = 49.7 bits (117), Expect = 2e-06
Identities = 41/123 (33%), Positives = 50/123 (40%)
Frame = +1
Query: 10 ASPSSEESAPASPDPSQQPPPQDPPSPAQSQPETSDDPPQQPPPQDPLQPEQSLSESPDP 69
A P+S SA A+P PS PP P +S P T+ PP PP P P + S P
Sbjct: 826 APPASSASA-ATPSPSSNKPP---PYIVKSSP-TASPPPPAPPAASPSAPPPLPTSSTPP 990
Query: 70 SQQSTLQDPLLLEQALNETAGSSQQPPQESSQSERSPPQSRRPEHQATESRLPAQQRPES 129
S S P TA +S P + S+ PP P A +S PA P S
Sbjct: 991 SPPSPAAPP--------STASTSPASPWAAPNSQSPPP----PSPPAVQSSTPAP*SPAS 1134
Query: 130 SQP 132
P
Sbjct: 1135PPP 1143
Score = 41.2 bits (95), Expect = 8e-04
Identities = 48/168 (28%), Positives = 64/168 (37%), Gaps = 6/168 (3%)
Frame = +1
Query: 11 SPSSEESAP--ASPDPSQQPPPQDPPSPAQSQP---ETSDDPPQQPPPQDPLQPEQSLSE 65
SPSS + P P+ PPP PP+ + S P TS PP P P P +
Sbjct: 862 SPSSNKPPPYIVKSSPTASPPPPAPPAASPSAPPPLPTSSTPPSPPSPAAPPSTASTSPA 1041
Query: 66 SPDPSQQSTLQDPLLLEQALNETAGSSQQPPQESSQSERSP-PQSRRPEHQATESRLPAQ 124
SP + S P + A S P S S P P S P +A + P
Sbjct: 1042 SPWAAPNSQSPPP------PSPPAVQSSTPAP*SPASPPPPTPPSAPPSGKACPNTPPPG 1203
Query: 125 QRPESSQPEHQATESRSLEQQQPPSLTPGRPASEFRLLRGSSSTSVPS 172
P S++ A +RS + +P FR +G S+ VPS
Sbjct: 1204 NSPYSTRVMISAA-TRSFLFPR*IFPSPAASRCSFR-RKGYSTWPVPS 1341
Score = 40.0 bits (92), Expect = 0.002
Identities = 30/97 (30%), Positives = 39/97 (39%), Gaps = 2/97 (2%)
Frame = +1
Query: 10 ASPSSEESAPASPDPSQQPPPQDPPSPAQSQPETSDDPP--QQPPPQDPLQPEQSLSESP 67
ASPS+ P S P P P PPS A + P + P Q PPP P + S +P
Sbjct: 943 ASPSAPPPLPTSSTPPSPPSPAAPPSTASTSPASPWAAPNSQSPPPPSPPAVQ---SSTP 1113
Query: 68 DPSQQSTLQDPLLLEQALNETAGSSQQPPQESSQSER 104
P ++ P + A + PP S S R
Sbjct: 1114 AP*SPASPPPPTPPSAPPSGKACPNTPPPGNSPYSTR 1224
Score = 33.9 bits (76), Expect = 0.12
Identities = 14/44 (31%), Positives = 21/44 (46%)
Frame = +1
Query: 9 PASPSSEESAPASPDPSQQPPPQDPPSPAQSQPETSDDPPQQPP 52
P+ P+ + S PA P+ PPP P +P + + PP P
Sbjct: 1081 PSPPAVQSSTPAP*SPASPPPPTPPSAPPSGKACPNTPPPGNSP 1212
>TC228611 weakly similar to GB|CAA72487.1|1890317|ATP26A peroxidase ATP26a
{Arabidopsis thaliana;} , partial (49%)
Length = 731
Score = 49.7 bits (117), Expect = 2e-06
Identities = 46/165 (27%), Positives = 60/165 (35%), Gaps = 1/165 (0%)
Frame = +3
Query: 13 SSEESAPASPDPSQQPPPQDPPSPAQSQPETSDDPPQ-QPPPQDPLQPEQSLSESPDPSQ 71
SS S S+ P P PP + S T+ P PP P +P SP PS
Sbjct: 123 SSARS*GTPSQASRSPAPPPPPPRSASSSTTASSPTAVTPPSSSPPRP------SPGPSA 284
Query: 72 QSTLQDPLLLEQALNETAGSSQQPPQESSQSERSPPQSRRPEHQATESRLPAQQRPESSQ 131
T P A T+ S+ +PP S SP + P AT S P P S
Sbjct: 285 TPTSTSP---SPATPSTSSSAPRPPLSSPAPTPSPAPTFSPPPPATSS--PCSAAPSSPS 449
Query: 132 PEHQATESRSLEQQQPPSLTPGRPASEFRLLRGSSSTSVPSLEST 176
AT PP+ +P P + S + + PS S+
Sbjct: 450 SSAAATAEPPSPPPYPPT-SPPPPCPSLKSPNSSPNAASPSKNSS 581
Score = 46.2 bits (108), Expect = 2e-05
Identities = 42/176 (23%), Positives = 67/176 (37%)
Frame = +3
Query: 10 ASPSSEESAPASPDPSQQPPPQDPPSPAQSQPETSDDPPQQPPPQDPLQPEQSLSESPDP 69
AS S+ S+P + P PP+ P P+ + TS P +P S S +P P
Sbjct: 198 ASSSTTASSPTAVTPPSSSPPRPSPGPSATPTSTSPSPATPSTSSSAPRPPLS-SPAPTP 374
Query: 70 SQQSTLQDPLLLEQALNETAGSSQQPPQESSQSERSPPQSRRPEHQATESRLPAQQRPES 129
S T P A + ++ P S+ + PP + P+ + P S
Sbjct: 375 SPAPTFSPP---PPATSSPCSAAPSSPSSSAAATAEPPSPPPYPPTSPPPPCPSLKSPNS 545
Query: 130 SQPEHQATESRSLEQQQPPSLTPGRPASEFRLLRGSSSTSVPSLESTRGGRAFARL 185
S +++ S PS + P+S +S T++P + R F RL
Sbjct: 546 SPNAASPSKNSSPSVGPTPSDSLTAPSSS-----RTSPTTLPPPTTLATLRGFKRL 698
Score = 43.1 bits (100), Expect = 2e-04
Identities = 37/143 (25%), Positives = 49/143 (33%)
Frame = +3
Query: 9 PASPSSEESAPASPDPSQQPPPQDPPSPAQSQPETSDDPPQQPPPQDPLQPEQSLSESPD 68
PA+PS+ SAP P S P P P+P S P + P P P + +E P
Sbjct: 309 PATPSTSSSAPRPPLSS--PAPTPSPAPTFSPPPPATSSPCSAAPSSPSSSAAATAEPPS 482
Query: 69 PSQQSTLQDPLLLEQALNETAGSSQQPPQESSQSERSPPQSRRPEHQATESRLPAQQRPE 128
P P PP S +S S P + P ++ S P
Sbjct: 483 PPPYPPTSPP----------------PPCPSLKSPNSSPNAASPSKNSSPS-----VGPT 599
Query: 129 SSQPEHQATESRSLEQQQPPSLT 151
S + SR+ PP T
Sbjct: 600 PSDSLTAPSSSRTSPTTLPPPTT 668
>TC216484 similar to UP|FLA1_ARATH (Q9FM65) Fasciclin-like arabinogalactan
protein 1 precursor, partial (58%)
Length = 1213
Score = 49.3 bits (116), Expect = 3e-06
Identities = 42/144 (29%), Positives = 56/144 (38%), Gaps = 1/144 (0%)
Frame = +1
Query: 9 PASPSSEESAPASPDPSQQPPPQDPPSPAQSQPETSDDPPQQPPPQDPLQPEQSLSESPD 68
P SP+ S PAS + PP SP+ + P S P P P ++ S S
Sbjct: 94 PPSPTRTTS-PASSQSTPSSPPSTTTSPSPTSPPKSTAKPPSPSAPSTTPPCRTFSRSTP 270
Query: 69 PSQQSTLQDPLLLEQALNETAGSSQQPPQESSQSERSPPQSRRPEHQATESRLPAQQRPE 128
PS S P T+ S+ P+ S++S +PP Q T+ P P
Sbjct: 271 PSTPSRTSSP--------STSSSTTSAPRSSTRSPTAPPS----PPQCTKPPAPPPDPPA 414
Query: 129 SS-QPEHQATESRSLEQQQPPSLT 151
SS P A +S S PP T
Sbjct: 415 SSTSPTSTAEKSHS-----PPKTT 471
Score = 40.0 bits (92), Expect = 0.002
Identities = 46/173 (26%), Positives = 65/173 (36%), Gaps = 18/173 (10%)
Frame = +1
Query: 3 PLDQDHPASPSSEESAPASPDPSQQPPPQDP--------PSPAQSQPETSDDPPQQPPPQ 54
P P SPS+ + P S+ PP P S S P +S P PP
Sbjct: 190 PKSTAKPPSPSAPSTTPPCRTFSRSTPPSTPSRTSSPSTSSSTTSAPRSSTRSPTAPP-- 363
Query: 55 DPLQPEQSLSESPDPSQQSTLQDPLLLEQALNETAGSSQQPPQESSQSERSPPQSRRPEH 114
P Q + + PDP ST E++ + ++ PQ SS + + +
Sbjct: 364 SPPQCTKPPAPPPDPPASST-SPTSTAEKSHSPPKTTTAHSPQRSSNP*KKSLTTSQ*SR 540
Query: 115 QATESRLPAQQRPESSQPEH-------QATESRS---LEQQQPPSLTPGRPAS 157
A S LP Q++P P + Q T++RS L P LTP S
Sbjct: 541 SAKFSPLP-QRKPLHPHPPNKTSPTLCQNTDARSSPTLSLLNPTRLTPSMTTS 696
Score = 28.5 bits (62), Expect = 5.1
Identities = 15/54 (27%), Positives = 25/54 (45%)
Frame = -3
Query: 91 SSQQPPQESSQSERSPPQSRRPEHQATESRLPAQQRPESSQPEHQATESRSLEQ 144
S QQ +E+ Q ++ P + +P ++ PAQ+R P SL+Q
Sbjct: 1187 SHQQHHRENQQEHQNHPPAHQPPSFSSSHSSPAQERRRQKXPXQXRXALPSLKQ 1026
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.309 0.128 0.363
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,368,776
Number of Sequences: 63676
Number of extensions: 371459
Number of successful extensions: 24374
Number of sequences better than 10.0: 1766
Number of HSP's better than 10.0 without gapping: 7061
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 14268
length of query: 373
length of database: 12,639,632
effective HSP length: 99
effective length of query: 274
effective length of database: 6,335,708
effective search space: 1735983992
effective search space used: 1735983992
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0120.7


