

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0058a.3
(579 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
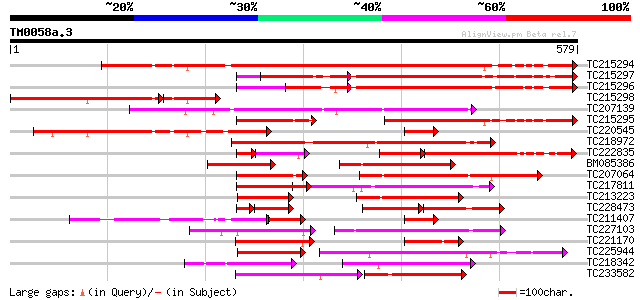
Score E
Sequences producing significant alignments: (bits) Value
TC215294 similar to UP|Q40090 (Q40090) SPF1 protein, partial (51%) 784 0.0
TC215297 similar to UP|Q40090 (Q40090) SPF1 protein, partial (33%) 431 e-121
TC215296 similar to UP|Q40090 (Q40090) SPF1 protein, partial (26%) 367 e-102
TC215298 similar to UP|Q40090 (Q40090) SPF1 protein, partial (21%) 248 6e-86
TC207139 similar to UP|Q8W1M6 (Q8W1M6) WRKY-like drought-induced... 293 1e-79
TC215295 similar to UP|Q94IB4 (Q94IB4) WRKY DNA-binding protein ... 292 3e-79
TC220545 similar to UP|Q40090 (Q40090) SPF1 protein, partial (23%) 291 4e-79
TC218972 similar to UP|Q9ZPL6 (Q9ZPL6) DNA-binding protein 2, pa... 268 4e-72
TC222835 similar to UP|Q6UN81 (Q6UN81) WRKY-type transcription f... 125 5e-49
BM085386 similar to GP|14530681|dbj WRKY DNA-binding protein {Ni... 181 8e-46
TC207064 similar to UP|Q8W1M6 (Q8W1M6) WRKY-like drought-induced... 177 1e-44
TC217811 similar to UP|Q39658 (Q39658) SPF1-like DNA-binding pro... 165 5e-41
TC213223 similar to UP|Q6VR10 (Q6VR10) WRKY 13 (Fragment), parti... 127 1e-29
TC228473 similar to GB|CAA63554.1|1064883|ATZAP1 ZAP1 {Arabidops... 77 2e-29
TC211407 similar to UP|Q39658 (Q39658) SPF1-like DNA-binding pro... 96 8e-29
TC227103 similar to UP|WR23_ARATH (O22900) Probable WRKY transcr... 122 4e-28
TC221170 similar to UP|Q94IB3 (Q94IB3) WRKY DNA-binding protein,... 121 7e-28
TC225944 similar to UP|Q6IET0 (Q6IET0) WRKY transcription factor... 119 5e-27
TC218342 similar to UP|Q6IEK9 (Q6IEK9) WRKY transcription factor... 113 2e-25
TC233582 similar to UP|WR32_ARATH (P59583) Probable WRKY transcr... 110 2e-24
>TC215294 similar to UP|Q40090 (Q40090) SPF1 protein, partial (51%)
Length = 1737
Score = 784 bits (2025), Expect = 0.0
Identities = 401/498 (80%), Positives = 425/498 (84%), Gaps = 12/498 (2%)
Frame = +1
Query: 94 LLNSSNILPSPTTGAFAAQSFNWKSSSGGNQQVVKEEDKSFSNFSFQTQQAPPPATTSTA 153
LLNSSNILPSPTTGAF AQSFNWKSSSGGNQQ+VKEEDKSFSNFSFQT+ PP +STA
Sbjct: 1 LLNSSNILPSPTTGAFVAQSFNWKSSSGGNQQIVKEEDKSFSNFSFQTRSGPP--ASSTA 174
Query: 154 TYQSSNVASQTQQQPWSYQEATKQDSM-----MMKTENSSSMQSFSPEIASVQTNHNNGF 208
TYQSSNV QTQQ PWS+QEATKQD+ MMKTENSSSMQSFSPEIASVQTNH+NGF
Sbjct: 175 TYQSSNVTVQTQQ-PWSFQEATKQDNFSSGKGMMKTENSSSMQSFSPEIASVQTNHSNGF 351
Query: 209 QSDYNNYQPQQQQLQPQTLNRRSDDGYNWRKYGQKQVKGSENPRSYYKCTYPNCPTKKKV 268
QSDY NY PQ Q TL+RRSDDGYNWRKYGQKQVKGSENPRSYYKCTYPNCPTKKKV
Sbjct: 352 QSDYGNYPPQSQ-----TLSRRSDDGYNWRKYGQKQVKGSENPRSYYKCTYPNCPTKKKV 516
Query: 269 ERSLDGQITEIVYKGTHNHPKPQATRRNSSSSSSLPIPHSNHISNDIQDQSYATHGSGQM 328
ERSLDGQITEIVYKGTHNHPKPQ TRRNSS+SSSL IPHSN I +I DQSYATHGSGQM
Sbjct: 517 ERSLDGQITEIVYKGTHNHPKPQNTRRNSSNSSSLAIPHSNSIRTEIPDQSYATHGSGQM 696
Query: 329 DSVATPENSSISIGDDDFEQSSQKCRSGGDELDEDEPDSKRWKVEGENEGISAPGSRTVR 388
DS ATPENSSISIGDDDFEQSSQKC+SGGDE DEDEPD+KRWK+EGENEG+SAPGSRTVR
Sbjct: 697 DSAATPENSSISIGDDDFEQSSQKCKSGGDEYDEDEPDAKRWKIEGENEGMSAPGSRTVR 876
Query: 389 EPRVVVQTTSDIDILDDGYRWRKYGQKVVKGNPNPRSYYKCTHPGCPVRKHVERASHDLR 448
EPRVVVQTTSDIDILDDGYRWRKYGQKVVKGNPNPRSYYKCTHPGCPVRKHVERASHDLR
Sbjct: 877 EPRVVVQTTSDIDILDDGYRWRKYGQKVVKGNPNPRSYYKCTHPGCPVRKHVERASHDLR 1056
Query: 449 AVITTYEGKHNHDVPAARGSGNHSVTRPLPNNTAN-------AIRPSSVTHQHHHNTNNN 501
AVITTYEGKHNHDVPAARGSG+HSV RP+PNN +N ++R V HQ ++N
Sbjct: 1057AVITTYEGKHNHDVPAARGSGSHSVNRPMPNNASNHTNTAATSVRLLPVIHQ-----SDN 1221
Query: 502 SLQSLRPQQAPDQVQSSPFTLEMLQQNQGGFGFSSGFGNSMGSYNMNNQQQQQLPDNVFS 561
SLQ+ R Q P+ SPFTLEML Q+ G FGF SGFGN M SY QQQL DNVFS
Sbjct: 1222SLQNQRSQAPPE--GQSPFTLEML-QSPGSFGF-SGFGNPMQSY----VNQQQLSDNVFS 1377
Query: 562 SRAKEEPRDDTFFDSLLC 579
SR KEEPRDD F +SLLC
Sbjct: 1378SRTKEEPRDDMFLESLLC 1431
>TC215297 similar to UP|Q40090 (Q40090) SPF1 protein, partial (33%)
Length = 1298
Score = 431 bits (1109), Expect = e-121
Identities = 232/325 (71%), Positives = 253/325 (77%), Gaps = 2/325 (0%)
Frame = +2
Query: 257 CTYPNCPTKKKVERSLDGQITEIVYKGTHNHPKPQATRRNSSSSSSLPIPHSNHISNDIQ 316
CTYPNCPTKKKVERSLDGQITEIVYKGTHNHPKPQA +RNS S+SSL IPHSNH I
Sbjct: 2 CTYPNCPTKKKVERSLDGQITEIVYKGTHNHPKPQAAKRNSLSASSLAIPHSNH--GGIN 175
Query: 317 DQSYATHGSGQMDSVATPENSSISIGDDDFEQSSQKCRSGGDELDEDEPDSKRWKVEGEN 376
+ + QMDSVATPENSSIS+ DDDF+ + +SGGDE D DEPD+KRW++EGEN
Sbjct: 176 ELPH------QMDSVATPENSSISMEDDDFDHT----KSGGDEFDNDEPDAKRWRIEGEN 325
Query: 377 EGISAPGSRTVREPRVVVQTTSDIDILDDGYRWRKYGQKVVKGNPNPRSYYKCTHPGCPV 436
EG+ A SRTVREPRVV QTTSDIDILDDGYRWRKYGQKVVKGNPNPRSYYKCT PGCPV
Sbjct: 326 EGMPAIESRTVREPRVVFQTTSDIDILDDGYRWRKYGQKVVKGNPNPRSYYKCTFPGCPV 505
Query: 437 RKHVERASHDLRAVITTYEGKHNHDVPAARGSGNHSVTRPLP-NNTANAIRPSSVTHQHH 495
RKHVERAS DLRAVITTYEGKHNHDVPAARGSGN+S+ R LP NT N S+ T +
Sbjct: 506 RKHVERASQDLRAVITTYEGKHNHDVPAARGSGNNSMNRSLPITNTTN--NTSAATSLY- 676
Query: 496 HNTNNNSLQSLRPQQAPDQVQSSPFTLEMLQQNQGGFGFSSGFGNS-MGSYNMNNQQQQQ 554
TNNNSLQSLRP AP++ SS F M Q + G FGF SGFGN MGSY NQQ
Sbjct: 677 --TNNNSLQSLRPPAAPERT-SSHFNPNMQQSSSGSFGF-SGFGNPLMGSY--TNQQS-- 832
Query: 555 LPDNVFSSRAKEEPRDDTFFDSLLC 579
DNVF +RAKEEP DD+F DS LC
Sbjct: 833 --DNVFITRAKEEPGDDSFLDSFLC 901
Score = 94.7 bits (234), Expect = 1e-19
Identities = 54/122 (44%), Positives = 73/122 (59%), Gaps = 3/122 (2%)
Frame = +2
Query: 232 DDGYNWRKYGQKQVKGSENPRSYYKCTYPNCPTKKKVER-SLDGQITEIVYKGTHNHPKP 290
DDGY WRKYGQK VKG+ NPRSYYKCT+P CP +K VER S D + Y+G HNH P
Sbjct: 407 DDGYRWRKYGQKVVKGNPNPRSYYKCTFPGCPVRKHVERASQDLRAVITTYEGKHNHDVP 586
Query: 291 QATRRNSSSSSSLPIPHSNHISNDIQDQSYATHGSG--QMDSVATPENSSISIGDDDFEQ 348
A R + ++S + +P +N +N S T+ + + A PE +S S + + +Q
Sbjct: 587 -AARGSGNNSMNRSLPITNTTNNTSAATSLYTNNNSLQSLRPPAAPERTS-SHFNPNMQQ 760
Query: 349 SS 350
SS
Sbjct: 761 SS 766
>TC215296 similar to UP|Q40090 (Q40090) SPF1 protein, partial (26%)
Length = 1278
Score = 367 bits (943), Expect = e-102
Identities = 202/301 (67%), Positives = 226/301 (74%), Gaps = 3/301 (0%)
Frame = +2
Query: 282 KGTHNHPKPQATRRNS-SSSSSLPIPHSNHISNDIQDQSYATHGSGQMDSVATPENSSIS 340
KGTHNHPKP + +RNS S+SSSL IPHSNH SN++ QMDSVATPENSSIS
Sbjct: 2 KGTHNHPKPSSAKRNSLSASSSLAIPHSNHGSNELPHH--------QMDSVATPENSSIS 157
Query: 341 IGDDDFEQSSQKCRSGGDELDEDEPDSKRWKVEGENEGISAPGSRTVREPRVVVQTTSDI 400
+ DDDF+ + +S E D DEPD+KRW++EGENEGISA GSRTVREPRVVVQTTSDI
Sbjct: 158 MDDDDFDHT----KSFLYEFDNDEPDAKRWRIEGENEGISAVGSRTVREPRVVVQTTSDI 325
Query: 401 DILDDGYRWRKYGQKVVKGNPNPRSYYKCTHPGCPVRKHVERASHDLRAVITTYEGKHNH 460
DILDDGYRWRKYGQKVVKGNPNPRSYYKCT PGCPVRKHVERAS DLRAVITTYEGKHNH
Sbjct: 326 DILDDGYRWRKYGQKVVKGNPNPRSYYKCTFPGCPVRKHVERASQDLRAVITTYEGKHNH 505
Query: 461 DVPAARGSGNHSVTRPLPNNTANAIRPSSVTHQHHHNTNNNSLQSLRPQQAPDQVQSSPF 520
DVPAARGSGN+S++R LP T +SV +TNNNSLQSLRP P++ S F
Sbjct: 506 DVPAARGSGNNSISRSLPIITNTTNNTTSVATS--ISTNNNSLQSLRPPAPPERPSLSHF 679
Query: 521 TLEMLQQNQGGFGFSSGFGNS-MGSYNMNNQQQQQLPDNVF-SSRAKEEPRDDTFFDSLL 578
M Q + G FGF SGFGN MGSY QQ +NVF ++R KEEP DD+F DSLL
Sbjct: 680 NPNM-QHSSGSFGF-SGFGNPLMGSY-----MNQQSYNNVFTTTRDKEEPGDDSFLDSLL 838
Query: 579 C 579
C
Sbjct: 839 C 841
Score = 96.3 bits (238), Expect = 3e-20
Identities = 56/124 (45%), Positives = 73/124 (58%), Gaps = 5/124 (4%)
Frame = +2
Query: 232 DDGYNWRKYGQKQVKGSENPRSYYKCTYPNCPTKKKVER-SLDGQITEIVYKGTHNHPKP 290
DDGY WRKYGQK VKG+ NPRSYYKCT+P CP +K VER S D + Y+G HNH P
Sbjct: 335 DDGYRWRKYGQKVVKGNPNPRSYYKCTFPGCPVRKHVERASQDLRAVITTYEGKHNHDVP 514
Query: 291 QAT-RRNSSSSSSLPIPHSNHISNDIQDQSYATHGSGQMDSV---ATPENSSISIGDDDF 346
A N+S S SLPI +N +N + + + + S+ A PE S+S + +
Sbjct: 515 AARGSGNNSISRSLPI-ITNTTNNTTSVATSISTNNNSLQSLRPPAPPERPSLSHFNPNM 691
Query: 347 EQSS 350
+ SS
Sbjct: 692 QHSS 703
>TC215298 similar to UP|Q40090 (Q40090) SPF1 protein, partial (21%)
Length = 701
Score = 248 bits (632), Expect(2) = 6e-86
Identities = 131/165 (79%), Positives = 138/165 (83%), Gaps = 8/165 (4%)
Frame = +3
Query: 1 MASSGGSLDTSAS--SFNNFTFSTHPFMTTSFSDLLASPISEDKPRGGLSDRIAERTGSG 58
MASS GSLDTSAS SF NFTFSTHPFMTTSFSDLLASP +KP G ++ERTGSG
Sbjct: 24 MASSSGSLDTSASANSFTNFTFSTHPFMTTSFSDLLASPTDNNKPPHGGGGGLSERTGSG 203
Query: 59 VPKFKSTPPPSLPLSPPTIF------IPPGLSPAELLDSPVLLNSSNILPSPTTGAFAAQ 112
VPKFKSTPPPSLPLSPP I IPPGLSPAELLDSPVLLNSSNILPSPTTGAF A+
Sbjct: 204 VPKFKSTPPPSLPLSPPPISPSSYFSIPPGLSPAELLDSPVLLNSSNILPSPTTGAFVAR 383
Query: 113 SFNWKSSSGGNQQVVKEEDKSFSNFSFQTQQAPPPATTSTATYQS 157
SFNWKSSSGGNQ++VKEEDK FSNFSFQTQQ PP +STATYQS
Sbjct: 384 SFNWKSSSGGNQRIVKEEDKGFSNFSFQTQQGPP--ASSTATYQS 512
Score = 89.0 bits (219), Expect(2) = 6e-86
Identities = 46/63 (73%), Positives = 49/63 (77%), Gaps = 5/63 (7%)
Frame = +1
Query: 158 SNVASQTQQQPWSYQEATKQDSM-----MMKTENSSSMQSFSPEIASVQTNHNNGFQSDY 212
SNV +QTQQ PWSY E TKQD+ MM TE SSSMQSFSPEIASVQ NH+NGFQSDY
Sbjct: 514 SNVTAQTQQ-PWSYHETTKQDNFSSGKSMMXTEKSSSMQSFSPEIASVQNNHSNGFQSDY 690
Query: 213 NNY 215
NY
Sbjct: 691 GNY 699
>TC207139 similar to UP|Q8W1M6 (Q8W1M6) WRKY-like drought-induced protein,
partial (85%)
Length = 1774
Score = 293 bits (750), Expect = 1e-79
Identities = 175/373 (46%), Positives = 222/373 (58%), Gaps = 19/373 (5%)
Frame = +2
Query: 123 NQQVVKEEDKSFSNFSFQTQQAPPPATTST-ATYQSSNVASQTQQQPWSYQEATKQD--- 178
N V + SF F + PA + + QS+ V + Q ++ + +
Sbjct: 83 NTNTVDDRKSSFFEFKPLNRSNKVPADFNNHVSKQSTQVEGPGKTQSFASSPLVESEIAV 262
Query: 179 -----SMMMKTENSSSMQSFSPEIASVQTNHNN----GFQSDYNNYQPQQQQLQPQTLNR 229
S+ + SS S ++ NH G Q+ + + + + +
Sbjct: 263 PSNELSLSSPVQKVSSSASAPVDVDLDDINHKGNTATGLQASHVEVRGSGLSVAAE---K 433
Query: 230 RSDDGYNWRKYGQKQVKGSENPRSYYKCTYPNCPTKKKVERSLDGQITEIVYKGTHNHPK 289
SDDGYNWRKYGQK VKGSE PRSYYKCT+PNC KK ERS DGQITEIVYKGTH+HPK
Sbjct: 434 TSDDGYNWRKYGQKLVKGSEFPRSYYKCTHPNCEVKKLFERSHDGQITEIVYKGTHDHPK 613
Query: 290 PQATRRNSSSS-SSLPIPHSNHISNDIQDQSYATHGSGQMDSVA----TPENSSISIGDD 344
PQ + R S+ + S+ S+ S +D AT GQ+ A TPE+S ++ DD
Sbjct: 614 PQPSCRYSTGTVMSIQGERSDKASMAGRDDK-ATAMYGQVSHAAEPNSTPESSPVATNDD 790
Query: 345 DFEQ-SSQKCRSGGDELDEDEPDSKRWKVEGENEGISAPGSRTVREPRVVVQTTSDIDIL 403
E + +E+D D+P SKR K+E N I+ P + +REPRVVVQT S++DIL
Sbjct: 791 GLEGVAGFVSNRTNEEVDNDDPFSKRRKMELGNVDIT-PVVKPIREPRVVVQTLSEVDIL 967
Query: 404 DDGYRWRKYGQKVVKGNPNPRSYYKCTHPGCPVRKHVERASHDLRAVITTYEGKHNHDVP 463
DDGYRWRKYGQKVV+GNPNPRSYYKCT+ GCPVRKHVERASHD +AVITTYEGKHNHDVP
Sbjct: 968 DDGYRWRKYGQKVVRGNPNPRSYYKCTNAGCPVRKHVERASHDPKAVITTYEGKHNHDVP 1147
Query: 464 AARGSGNHSVTRP 476
AAR S +H + P
Sbjct: 1148AARNS-SHDMAVP 1183
>TC215295 similar to UP|Q94IB4 (Q94IB4) WRKY DNA-binding protein (Fragment),
partial (34%)
Length = 925
Score = 292 bits (747), Expect = 3e-79
Identities = 156/204 (76%), Positives = 165/204 (80%), Gaps = 7/204 (3%)
Frame = +1
Query: 383 GSRTVREPRVVVQTTSDIDILDDGYRWRKYGQKVVKGNPNPRSYYKCTHPGCPVRKHVER 442
GSRTVREPRVVVQTTSDIDILDDGYRWRKYGQKVVKGNPNPRSYYKCTHPGCPVRKHVER
Sbjct: 1 GSRTVREPRVVVQTTSDIDILDDGYRWRKYGQKVVKGNPNPRSYYKCTHPGCPVRKHVER 180
Query: 443 ASHDLRAVITTYEGKHNHDVPAARGSGNHSVTRPLPNNTAN-------AIRPSSVTHQHH 495
ASHDLRAVITTYEGKHNHDVPAARGSG+HSV RP+PNN +N AI P V QH
Sbjct: 181 ASHDLRAVITTYEGKHNHDVPAARGSGSHSVNRPMPNNASNPTNTAATAISPLQVI-QHS 357
Query: 496 HNTNNNSLQSLRPQQAPDQVQSSPFTLEMLQQNQGGFGFSSGFGNSMGSYNMNNQQQQQL 555
N++ N + QAP + Q SPFTLEML Q+ G FGF SGFGN M SY NQQQQQL
Sbjct: 358 DNSHQN-----QRSQAPPEGQ-SPFTLEML-QSPGSFGF-SGFGNPMQSY--MNQQQQQL 507
Query: 556 PDNVFSSRAKEEPRDDTFFDSLLC 579
DNVFSSRAKEEPRDD F +SLLC
Sbjct: 508 SDNVFSSRAKEEPRDDMFLESLLC 579
Score = 91.7 bits (226), Expect = 8e-19
Identities = 47/83 (56%), Positives = 56/83 (66%), Gaps = 1/83 (1%)
Frame = +1
Query: 232 DDGYNWRKYGQKQVKGSENPRSYYKCTYPNCPTKKKVER-SLDGQITEIVYKGTHNHPKP 290
DDGY WRKYGQK VKG+ NPRSYYKCT+P CP +K VER S D + Y+G HNH P
Sbjct: 64 DDGYRWRKYGQKVVKGNPNPRSYYKCTHPGCPVRKHVERASHDLRAVITTYEGKHNHDVP 243
Query: 291 QATRRNSSSSSSLPIPHSNHISN 313
A R + S S + P+P N+ SN
Sbjct: 244 -AARGSGSHSVNRPMP--NNASN 303
>TC220545 similar to UP|Q40090 (Q40090) SPF1 protein, partial (23%)
Length = 785
Score = 291 bits (746), Expect = 4e-79
Identities = 167/267 (62%), Positives = 186/267 (69%), Gaps = 24/267 (8%)
Frame = +1
Query: 25 FMTTSFSDLLASPISEDK----PRGGLSDRIAERTGS--GVPKFKSTPPPSLPLSP--PT 76
FM TSFSDLL+SP ++ R L +++ GS GVPKFKSTPPPSLPLS P
Sbjct: 22 FMITSFSDLLSSPTTDHNLGVSERPHLKSSSSDQAGSSGGVPKFKSTPPPSLPLSHHLPP 201
Query: 77 IF---------IPPGLSPAELLDSPVLLNSSNILPSPTTGAFAAQSFNWKSSSGGNQQVV 127
IF IP GLS AELLDSPVLLNSSN+LPSPTTG+FA Q FNWKSS G +QQ V
Sbjct: 202 IFSPSSYFNFNIPHGLSLAELLDSPVLLNSSNVLPSPTTGSFAGQGFNWKSSYGESQQHV 381
Query: 128 KEEDKSFSNFSFQTQQAPPPATTSTATYQSSNVASQTQQQPWSYQEATKQDSM------- 180
KEEDKSFS+FSFQTQ PP S+ +QSS QT WS+ E KQD
Sbjct: 382 KEEDKSFSSFSFQTQTHPP--LPSSNGFQSSTGIVQT---GWSFPETAKQDGFASRISMS 546
Query: 181 MMKTENSSSMQSFSPEIASVQTNHNNGFQSDYNNYQPQQQQLQPQTLNRRSDDGYNWRKY 240
M+KTE +S+MQS +PE NH NGFQSD+ NYQPQ Q QTL+RRSDDGYNWRKY
Sbjct: 547 MVKTETTSAMQSLTPE----NNNHRNGFQSDHKNYQPQ----QVQTLSRRSDDGYNWRKY 702
Query: 241 GQKQVKGSENPRSYYKCTYPNCPTKKK 267
GQKQVKGSENPRSYYKCTYPNCPTKKK
Sbjct: 703 GQKQVKGSENPRSYYKCTYPNCPTKKK 783
Score = 69.7 bits (169), Expect = 3e-12
Identities = 27/35 (77%), Positives = 30/35 (85%)
Frame = +1
Query: 404 DDGYRWRKYGQKVVKGNPNPRSYYKCTHPGCPVRK 438
DDGY WRKYGQK VKG+ NPRSYYKCT+P CP +K
Sbjct: 676 DDGYNWRKYGQKQVKGSENPRSYYKCTYPNCPTKK 780
>TC218972 similar to UP|Q9ZPL6 (Q9ZPL6) DNA-binding protein 2, partial (33%)
Length = 1177
Score = 268 bits (686), Expect = 4e-72
Identities = 140/274 (51%), Positives = 184/274 (67%), Gaps = 4/274 (1%)
Frame = +2
Query: 227 LNRRSDDGYNWRKYGQKQVKGSENPRSYYKCTYPNCPTKKKVERSLDGQITEIVYKGTHN 286
+++ +DDGYNWRKYGQKQVKGSE PRSYYKCT+ NC KKKVERSL+G +T I+YKG HN
Sbjct: 26 VDKPADDGYNWRKYGQKQVKGSEFPRSYYKCTHSNCSVKKKVERSLEGHVTAIIYKGEHN 205
Query: 287 HPKPQATRRNSSSSSSLPIPHSNHISNDIQDQSYATHGSGQMDSVATPENSSISIGDDDF 346
H +P +R+ + +S + +++IQ +T+ +S++ + S D
Sbjct: 206 HQRPHPNKRSKDTMTS-------NANSNIQGSVDSTYQGTTTNSMSKMDPESSQATADHL 364
Query: 347 EQSSQKCRSGGDELDEDE----PDSKRWKVEGENEGISAPGSRTVREPRVVVQTTSDIDI 402
+S+ G E + DE PD KR K E ++ A RTV EPR++VQTTS++D+
Sbjct: 365 SGTSESEEVGDHETEVDEKNVEPDPKRRKAE-VSQSDPASSHRTVTEPRIIVQTTSEVDL 541
Query: 403 LDDGYRWRKYGQKVVKGNPNPRSYYKCTHPGCPVRKHVERASHDLRAVITTYEGKHNHDV 462
LDDGYRWRKYGQKVVKGNP RSYYKCT GC VRKHVERAS D +AVITTYEGKHNHDV
Sbjct: 542 LDDGYRWRKYGQKVVKGNPYXRSYYKCTTQGCNVRKHVERASTDPKAVITTYEGKHNHDV 721
Query: 463 PAARGSGNHSVTRPLPNNTANAIRPSSVTHQHHH 496
PAA+ N+S T + +NTA+ ++ + + H+
Sbjct: 722 PAAK---NNSHT--MASNTASQLKSHNTNPEKHN 808
>TC222835 similar to UP|Q6UN81 (Q6UN81) WRKY-type transcription factor,
partial (18%)
Length = 714
Score = 125 bits (314), Expect(2) = 5e-49
Identities = 76/156 (48%), Positives = 99/156 (62%), Gaps = 1/156 (0%)
Frame = +3
Query: 424 RSYYKCTHPGCPVRKHVERASHDLRAVITTYEGKHNHDVPAARGSGNHSVTRPLPNNTAN 483
RSYYKCT PGC VRKHVERA+HD++AVITTYEGKHNHDVPAARGSGN+ + R N++
Sbjct: 243 RSYYKCTAPGCSVRKHVERAAHDIKAVITTYEGKHNHDVPAARGSGNYYMNRNSLNSSIP 422
Query: 484 A-IRPSSVTHQHHHNTNNNSLQSLRPQQAPDQVQSSPFTLEMLQQNQGGFGFSSGFGNSM 542
A IRPS+V + ++ NSL + A +S +L+ +N G FG+S+ SM
Sbjct: 423 APIRPSAVNCYSNSSSFTNSLYNNTRHPATGNQESC--SLDKF-KNPGSFGYSA-LNRSM 590
Query: 543 GSYNMNNQQQQQLPDNVFSSRAKEEPRDDTFFDSLL 578
S+ Q Q D S+AK+E +DD+F S L
Sbjct: 591 SSHT----DQAQYTD-AAHSKAKDERKDDSFLQSFL 683
Score = 88.2 bits (217), Expect(2) = 5e-49
Identities = 41/47 (87%), Positives = 43/47 (91%)
Frame = +1
Query: 378 GISAPGSRTVREPRVVVQTTSDIDILDDGYRWRKYGQKVVKGNPNPR 424
G S GSRTVREPRVVVQTTS+IDILDDG+RWRKYGQKVVKGNPN R
Sbjct: 1 GYSVSGSRTVREPRVVVQTTSEIDILDDGFRWRKYGQKVVKGNPNAR 141
Score = 47.4 bits (111), Expect(2) = 5e-11
Identities = 26/62 (41%), Positives = 33/62 (52%), Gaps = 7/62 (11%)
Frame = +3
Query: 252 RSYYKCTYPNCPTKKKVERSL-DGQITEIVYKGTHNHPKPQAT------RRNSSSSSSLP 304
RSYYKCT P C +K VER+ D + Y+G HNH P A +S +SS+P
Sbjct: 243 RSYYKCTAPGCSVRKHVERAAHDIKAVITTYEGKHNHDVPAARGSGNYYMNRNSLNSSIP 422
Query: 305 IP 306
P
Sbjct: 423 AP 428
Score = 38.5 bits (88), Expect(2) = 5e-11
Identities = 15/21 (71%), Positives = 17/21 (80%)
Frame = +1
Query: 232 DDGYNWRKYGQKQVKGSENPR 252
DDG+ WRKYGQK VKG+ N R
Sbjct: 79 DDGFRWRKYGQKVVKGNPNAR 141
>BM085386 similar to GP|14530681|dbj WRKY DNA-binding protein {Nicotiana
tabacum}, partial (17%)
Length = 410
Score = 181 bits (459), Expect = 8e-46
Identities = 88/121 (72%), Positives = 102/121 (83%), Gaps = 2/121 (1%)
Frame = +1
Query: 337 SSISIGDDDFEQSSQKCRSGGDELDEDEPDSKRWKVEGENEGIS--APGSRTVREPRVVV 394
S S+GD+D EQ+SQ SGG + D+ ++KRWK E EN+G S + GSRTV+EP+VVV
Sbjct: 49 SDQSVGDEDLEQTSQTSYSGGGD-DDLGNEAKRWKGENENDGYSYSSAGSRTVKEPKVVV 225
Query: 395 QTTSDIDILDDGYRWRKYGQKVVKGNPNPRSYYKCTHPGCPVRKHVERASHDLRAVITTY 454
QTTS+IDILDDGYRWRKYGQKVVKGNPNPRSYYKC PGCPVRKHVERASHD++AVITTY
Sbjct: 226 QTTSEIDILDDGYRWRKYGQKVVKGNPNPRSYYKCVAPGCPVRKHVERASHDMKAVITTY 405
Query: 455 E 455
E
Sbjct: 406 E 408
Score = 72.4 bits (176), Expect = 5e-13
Identities = 35/72 (48%), Positives = 46/72 (63%), Gaps = 3/72 (4%)
Frame = +1
Query: 203 NHNNGFQ-SDYNNYQPQQQQLQPQTLNRRS--DDGYNWRKYGQKQVKGSENPRSYYKCTY 259
N N+G+ S + ++ ++ QT + DDGY WRKYGQK VKG+ NPRSYYKC
Sbjct: 157 NENDGYSYSSAGSRTVKEPKVVVQTTSEIDILDDGYRWRKYGQKVVKGNPNPRSYYKCVA 336
Query: 260 PNCPTKKKVERS 271
P CP +K VER+
Sbjct: 337 PGCPVRKHVERA 372
>TC207064 similar to UP|Q8W1M6 (Q8W1M6) WRKY-like drought-induced protein,
partial (42%)
Length = 1782
Score = 177 bits (449), Expect = 1e-44
Identities = 97/199 (48%), Positives = 123/199 (61%), Gaps = 12/199 (6%)
Frame = +3
Query: 358 DELDEDEPDSKRWKVEGENEGISAPGSRTVREPRVVVQTTSDIDILDDGYRWRKYGQKVV 417
DE+D+D+P SKR K++ I+ P + +REPRVVVQT S++DILDDGYRWRKYGQKVV
Sbjct: 51 DEVDDDDPFSKRRKMDVGIADIT-PVVKPIREPRVVVQTLSEVDILDDGYRWRKYGQKVV 227
Query: 418 KGNPNPRSYYKCTHPGCPVRKHVERASHDLRAVITTYEGKHNHDVPAARGSGNHSVTRPL 477
+GNPNPRSYYKCT+ GCPVRKHVERASHD +AVITTYEGKHNHDVP AR S H + P
Sbjct: 228 RGNPNPRSYYKCTNTGCPVRKHVERASHDPKAVITTYEGKHNHDVPTARNS-CHDMAGPA 404
Query: 478 PNNTANAIRPSSV----------THQHHHNTNNNSLQSLRPQQAPDQVQSSPFTLEMLQQ 527
+ +RP NT+N+ + + + Q+ +S + +
Sbjct: 405 SASGQTRVRPEESDTISLDLGMGISPAAENTSNSQGRMMLSEFGDSQIHTSNSNFKFVHT 584
Query: 528 N--QGGFGFSSGFGNSMGS 544
G FG + N GS
Sbjct: 585 TTAPGYFGVLNNNSNPYGS 641
Score = 82.4 bits (202), Expect = 5e-16
Identities = 41/74 (55%), Positives = 47/74 (63%), Gaps = 1/74 (1%)
Frame = +3
Query: 232 DDGYNWRKYGQKQVKGSENPRSYYKCTYPNCPTKKKVER-SLDGQITEIVYKGTHNHPKP 290
DDGY WRKYGQK V+G+ NPRSYYKCT CP +K VER S D + Y+G HNH P
Sbjct: 186 DDGYRWRKYGQKVVRGNPNPRSYYKCTNTGCPVRKHVERASHDPKAVITTYEGKHNHDVP 365
Query: 291 QATRRNSSSSSSLP 304
T RNS + P
Sbjct: 366 --TARNSCHDMAGP 401
>TC217811 similar to UP|Q39658 (Q39658) SPF1-like DNA-binding protein,
partial (28%)
Length = 1010
Score = 165 bits (418), Expect = 5e-41
Identities = 96/224 (42%), Positives = 134/224 (58%), Gaps = 17/224 (7%)
Frame = +2
Query: 289 KPQATRRNSSSSSSLPIPHSNHISNDIQDQSYATHGSGQMDSVATP-ENSSISIGDDDFE 347
KPQA RR +S S + +++ + +S + GQ++ ++ NSS+ D
Sbjct: 5 KPQANRRAKDNSDS-----NGNVTVQPKSESNSQGWVGQLNKLSEKIPNSSVPKSDQTSN 169
Query: 348 QS---------SQKCRSGG-----DELDEDEPDSKRWKVEGENEGISAP--GSRTVREPR 391
Q S + G +E D+ EP+ KR + G+S +TV EP+
Sbjct: 170 QGAPPRQLLPGSNESEEVGNVDNREEADDGEPNPKR---RNTDVGVSEVPLSQKTVTEPK 340
Query: 392 VVVQTTSDIDILDDGYRWRKYGQKVVKGNPNPRSYYKCTHPGCPVRKHVERASHDLRAVI 451
++VQT S++D+LDDGYRWRKYGQKVVKGNP+PRSYYKCT GC VRKHVERAS D +AVI
Sbjct: 341 IIVQTRSEVDLLDDGYRWRKYGQKVVKGNPHPRSYYKCTSAGCNVRKHVERASMDPKAVI 520
Query: 452 TTYEGKHNHDVPAARGSGNHSVTRPLPNNTANAIRPSSVTHQHH 495
TTYEGKHNHDVPAAR S +++ ++ + ++P +V + H
Sbjct: 521 TTYEGKHNHDVPAARNSSHNTA-----SSNSMPLKPHNVVPEKH 637
Score = 87.4 bits (215), Expect = 2e-17
Identities = 43/81 (53%), Positives = 56/81 (69%), Gaps = 4/81 (4%)
Frame = +2
Query: 232 DDGYNWRKYGQKQVKGSENPRSYYKCTYPNCPTKKKVER-SLDGQITEIVYKGTHNHPKP 290
DDGY WRKYGQK VKG+ +PRSYYKCT C +K VER S+D + Y+G HNH P
Sbjct: 377 DDGYRWRKYGQKVVKGNPHPRSYYKCTSAGCNVRKHVERASMDPKAVITTYEGKHNHDVP 556
Query: 291 QA--TRRNSSSSSSLPI-PHS 308
A + N++SS+S+P+ PH+
Sbjct: 557 AARNSSHNTASSNSMPLKPHN 619
>TC213223 similar to UP|Q6VR10 (Q6VR10) WRKY 13 (Fragment), partial (36%)
Length = 416
Score = 127 bits (319), Expect = 1e-29
Identities = 62/109 (56%), Positives = 80/109 (72%)
Frame = +3
Query: 355 SGGDELDEDEPDSKRWKVEGENEGISAPGSRTVREPRVVVQTTSDIDILDDGYRWRKYGQ 414
S G E EDE SKR K E ++ +A + EPR+V+Q+ +D ++L DG+RWRKYGQ
Sbjct: 87 SKGFEAQEDEHRSKRRKNENQSNE-AALSEEGLVEPRIVMQSFTDSEVLGDGFRWRKYGQ 263
Query: 415 KVVKGNPNPRSYYKCTHPGCPVRKHVERASHDLRAVITTYEGKHNHDVP 463
KVVKGNP PRSY++CT+ C VRKHVERA D R+ +TTYEGKHNH++P
Sbjct: 264 KVVKGNPYPRSYFRCTNIMCNVRKHVERAIDDPRSCVTTYEGKHNHEMP 410
Score = 67.8 bits (164), Expect = 1e-11
Identities = 30/59 (50%), Positives = 40/59 (66%), Gaps = 1/59 (1%)
Frame = +3
Query: 233 DGYNWRKYGQKQVKGSENPRSYYKCTYPNCPTKKKVERSLDGQITEI-VYKGTHNHPKP 290
DG+ WRKYGQK VKG+ PRSY++CT C +K VER++D + + Y+G HNH P
Sbjct: 234 DGFRWRKYGQKVVKGNPYPRSYFRCTNIMCNVRKHVERAIDDPRSCVTTYEGKHNHEMP 410
>TC228473 similar to GB|CAA63554.1|1064883|ATZAP1 ZAP1 {Arabidopsis
thaliana;} , partial (16%)
Length = 1173
Score = 77.0 bits (188), Expect(2) = 2e-29
Identities = 36/83 (43%), Positives = 52/83 (62%)
Frame = +3
Query: 423 PRSYYKCTHPGCPVRKHVERASHDLRAVITTYEGKHNHDVPAARGSGNHSVTRPLPNNTA 482
P SYY+C++PGCPV+KHVERAS+D + VITTYEG+H+H++P R +VT+ NT
Sbjct: 219 P*SYYRCSNPGCPVKKHVERASYDSKTVITTYEGQHDHEIPPGR-----TVTQNAATNTR 383
Query: 483 NAIRPSSVTHQHHHNTNNNSLQS 505
+ NT++ +S
Sbjct: 384 TTATNGKAGTKSEGNTDDTGERS 452
Score = 70.9 bits (172), Expect(2) = 2e-29
Identities = 32/62 (51%), Positives = 43/62 (68%)
Frame = +1
Query: 361 DEDEPDSKRWKVEGENEGISAPGSRTVREPRVVVQTTSDIDILDDGYRWRKYGQKVVKGN 420
D EP+SKR K + N ++ T VVVQT+S++D+++DGYRWRKYGQK+VKGN
Sbjct: 31 DNKEPESKRLKKDNSNADVARVDMSTRESRVVVVQTSSEVDLVNDGYRWRKYGQKLVKGN 210
Query: 421 PN 422
N
Sbjct: 211 TN 216
Score = 44.3 bits (103), Expect(2) = 1e-09
Identities = 20/41 (48%), Positives = 25/41 (60%), Gaps = 1/41 (2%)
Frame = +3
Query: 251 PRSYYKCTYPNCPTKKKVER-SLDGQITEIVYKGTHNHPKP 290
P SYY+C+ P CP KK VER S D + Y+G H+H P
Sbjct: 219 P*SYYRCSNPGCPVKKHVERASYDSKTVITTYEGQHDHEIP 341
Score = 36.6 bits (83), Expect(2) = 1e-09
Identities = 14/19 (73%), Positives = 16/19 (83%)
Frame = +1
Query: 232 DDGYNWRKYGQKQVKGSEN 250
+DGY WRKYGQK VKG+ N
Sbjct: 160 NDGYRWRKYGQKLVKGNTN 216
>TC211407 similar to UP|Q39658 (Q39658) SPF1-like DNA-binding protein,
partial (33%)
Length = 784
Score = 95.9 bits (237), Expect(2) = 8e-29
Identities = 70/209 (33%), Positives = 99/209 (46%), Gaps = 3/209 (1%)
Frame = +2
Query: 62 FKSTPPPSLPLSPPTIF-IPPGLSPAELLDSPVLLNSSNILPSPTTGAFAAQSFNWKSSS 120
FK + P +L ++ +F +PPGLSP+ L+SP G F+ QS S
Sbjct: 77 FKQSRPMNLVIARSPVFTVPPGLSPSGFLNSP--------------GFFSPQSPFGMSHQ 214
Query: 121 GGNQQVVKEEDKSFSNFSFQTQQAPPPATTSTATYQSSNVASQTQQQPWSYQEATKQD-- 178
QV + + S+ Q P T T +Q ++ EA++Q
Sbjct: 215 QALAQVTAQAVLAQSHMHMQADYQMPAVTAPTEP--------PVRQLSFALNEASEQQVV 370
Query: 179 SMMMKTENSSSMQSFSPEIASVQTNHNNGFQSDYNNYQPQQQQLQPQTLNRRSDDGYNWR 238
S + + Q +PE++ Q+D YQP Q +++ +DDGYNWR
Sbjct: 371 SCVSSVSEPRNAQLEAPELS----------QAD-KKYQPSSQ-----AIDKPADDGYNWR 502
Query: 239 KYGQKQVKGSENPRSYYKCTYPNCPTKKK 267
KYGQKQVKGSE PRSYYKCT+ NC K+K
Sbjct: 503 KYGQKQVKGSEYPRSYYKCTHLNCVVKEK 589
Score = 61.6 bits (148), Expect = 9e-10
Identities = 25/35 (71%), Positives = 28/35 (79%)
Frame = +2
Query: 404 DDGYRWRKYGQKVVKGNPNPRSYYKCTHPGCPVRK 438
DDGY WRKYGQK VKG+ PRSYYKCTH C V++
Sbjct: 482 DDGYNWRKYGQKQVKGSEYPRSYYKCTHLNCVVKE 586
Score = 50.1 bits (118), Expect(2) = 8e-29
Identities = 23/38 (60%), Positives = 26/38 (67%)
Frame = +3
Query: 265 KKKVERSLDGQITEIVYKGTHNHPKPQATRRNSSSSSS 302
KKKVER+ DG ITEI+YKG HNH K A RR + S
Sbjct: 582 KKKVERAPDGHITEIIYKGQHNHEKAXANRRAQDNXDS 695
>TC227103 similar to UP|WR23_ARATH (O22900) Probable WRKY transcription
factor 23 (WRKY DNA-binding protein 23), partial (36%)
Length = 980
Score = 122 bits (306), Expect = 4e-28
Identities = 67/175 (38%), Positives = 93/175 (52%)
Frame = +2
Query: 332 ATPENSSISIGDDDFEQSSQKCRSGGDELDEDEPDSKRWKVEGENEGISAPGSRTVREPR 391
ATP +SSIS D Q ++ D+ +ED+ + + + + + REPR
Sbjct: 437 ATPNSSSISSASSDAVNDEQN-KTLLDQAEEDDDEEEGQQKTKKQLKPKKTNQKRQREPR 613
Query: 392 VVVQTTSDIDILDDGYRWRKYGQKVVKGNPNPRSYYKCTHPGCPVRKHVERASHDLRAVI 451
T S++D L+DGYRWRKYGQK VK +P PRSYY+CT C V+K VER+ D V+
Sbjct: 614 FAFMTKSEVDHLEDGYRWRKYGQKAVKNSPFPRSYYRCTSVSCNVKKRVERSFTDPSVVV 793
Query: 452 TTYEGKHNHDVPAARGSGNHSVTRPLPNNTANAIRPSSVTHQHHHNTNNNSLQSL 506
TTYEG+H H P SG V+ NN + + P + Q+ H + Q L
Sbjct: 794 TTYEGQHTHPSPVMPRSG---VSAGYANNFGSVLPPGNYLSQYQHYHHQQQQQHL 949
Score = 84.3 bits (207), Expect = 1e-16
Identities = 53/156 (33%), Positives = 78/156 (49%), Gaps = 27/156 (17%)
Frame = +2
Query: 184 TENSSSMQSFSPEIASVQTNHNNGFQSDYNN-----YQPQQQQLQPQTLNRRS------- 231
T NSSS+ S S + + + N Q++ ++ Q ++QL+P+ N++
Sbjct: 440 TPNSSSISSASSDAVNDEQNKTLLDQAEEDDDEEEGQQKTKKQLKPKKTNQKRQREPRFA 619
Query: 232 ----------DDGYNWRKYGQKQVKGSENPRSYYKCTYPNCPTKKKVERSL-DGQITEIV 280
+DGY WRKYGQK VK S PRSYY+CT +C KK+VERS D +
Sbjct: 620 FMTKSEVDHLEDGYRWRKYGQKAVKNSPFPRSYYRCTSVSCNVKKRVERSFTDPSVVVTT 799
Query: 281 YKGTHNHPKPQATRRNSS----SSSSLPIPHSNHIS 312
Y+G H HP P R S ++ +P N++S
Sbjct: 800 YEGQHTHPSPVMPRSGVSAGYANNFGSVLPPGNYLS 907
>TC221170 similar to UP|Q94IB3 (Q94IB3) WRKY DNA-binding protein, partial
(14%)
Length = 936
Score = 121 bits (304), Expect = 7e-28
Identities = 54/85 (63%), Positives = 64/85 (74%), Gaps = 4/85 (4%)
Frame = +3
Query: 231 SDDGYNWRKYGQKQVKGSENPRSYYKCTYPNCPTKKKVERSLDGQITEIVYKGTHNHPKP 290
S+DGYNWRKYGQKQVKGSE P SYYKCT+PNC KKKVERS +G ITEI+YKGTHNHPKP
Sbjct: 426 SEDGYNWRKYGQKQVKGSEYPLSYYKCTHPNCQVKKKVERSHEGHITEIIYKGTHNHPKP 605
Query: 291 QATRRNS----SSSSSLPIPHSNHI 311
RR+ + + + + H H+
Sbjct: 606 PPNRRSGIGLVNLHTDMQVDHPEHV 680
Score = 80.1 bits (196), Expect = 3e-15
Identities = 36/60 (60%), Positives = 41/60 (68%)
Frame = +3
Query: 404 DDGYRWRKYGQKVVKGNPNPRSYYKCTHPGCPVRKHVERASHDLRAVITTYEGKHNHDVP 463
+DGY WRKYGQK VKG+ P SYYKCTHP C V+K VER SH+ Y+G HNH P
Sbjct: 429 EDGYNWRKYGQKQVKGSEYPLSYYKCTHPNCQVKKKVER-SHEGHITEIIYKGTHNHPKP 605
>TC225944 similar to UP|Q6IET0 (Q6IET0) WRKY transcription factor 1, partial
(41%)
Length = 1504
Score = 119 bits (297), Expect = 5e-27
Identities = 95/290 (32%), Positives = 132/290 (44%), Gaps = 37/290 (12%)
Frame = +1
Query: 317 DQSYATHGSGQMDSVATPENSSISIGDDDFEQSSQ-KCRSGGDELDEDEPD----SKRWK 371
D + S + +TP+N +I G D +++ K + G +E + E +K K
Sbjct: 238 DDQVSDSSSDERTRSSTPQNHNIEAGARDGARNNNGKSQLGREESPDSESQGWGPNKLQK 417
Query: 372 VEGENEGISAPGSRTVREPRVVVQTTSDIDILDDGYRWRKYGQKVVKGNPNPRSYYKCTH 431
V N + T+R+ RV V+ S+ ++ DG +WRKYGQK+ KGNP PR+YY+CT
Sbjct: 418 VNPSNPMDQSTAEATMRKARVSVRARSEAPMISDGCQWRKYGQKMAKGNPCPRAYYRCTM 597
Query: 432 P-GCPVRKHVERASHDLRAVITTYEGKHNHDVPAA----------------RGSGNHSVT 474
GCPVRK V+R + D ++TTYEG HNH +P A GS + +
Sbjct: 598 AVGCPVRKQVQRCADDRTILVTTYEGTHNHPLPPAAMAMASTTAAAATMLLSGSMSSADG 777
Query: 475 RPLPNNTANAIRPSS--------------VTHQHHHNTNNNSLQSLRPQQAPDQVQSSPF 520
PN A AI P S VT HN N LQ RP +PF
Sbjct: 778 VMNPNLLARAILPCSTSMATLSASAPFPTVTLDLTHNP--NPLQFQRP--------GAPF 927
Query: 521 TLEMLQQNQGGFGFSSGFGNSMGSYNMNNQQQQQLPDNVFSSR-AKEEPR 569
+ LQ FG S + YN + QL +V SS+ A + PR
Sbjct: 928 QVPFLQAQPQNFG-SGATPIAQALYNQSKFSGLQLSQDVGSSQLAPQAPR 1074
Score = 73.6 bits (179), Expect = 2e-13
Identities = 34/72 (47%), Positives = 47/72 (65%), Gaps = 2/72 (2%)
Frame = +1
Query: 233 DGYNWRKYGQKQVKGSENPRSYYKCTYP-NCPTKKKVERSLDGQ-ITEIVYKGTHNHPKP 290
DG WRKYGQK KG+ PR+YY+CT CP +K+V+R D + I Y+GTHNHP P
Sbjct: 517 DGCQWRKYGQKMAKGNPCPRAYYRCTMAVGCPVRKQVQRCADDRTILVTTYEGTHNHPLP 696
Query: 291 QATRRNSSSSSS 302
A +S++++
Sbjct: 697 PAAMAMASTTAA 732
>TC218342 similar to UP|Q6IEK9 (Q6IEK9) WRKY transcription factor 72, partial
(43%)
Length = 775
Score = 113 bits (283), Expect = 2e-25
Identities = 58/141 (41%), Positives = 76/141 (53%), Gaps = 6/141 (4%)
Frame = +1
Query: 341 IGDDDFEQSSQKCRSG-GDELDEDEPDSKRWKVEGE-----NEGISAPGSRTVREPRVVV 394
+G DD + + G G+ L V E N G + V++PR
Sbjct: 40 LGSDDHDNGGEGGGDGDGNMLMSQISGGSNTNVSDELGGSGNSNNKKKGEKKVKKPRYAF 219
Query: 395 QTTSDIDILDDGYRWRKYGQKVVKGNPNPRSYYKCTHPGCPVRKHVERASHDLRAVITTY 454
QT S +DILDDGYRWRKYGQK VK N PRSYY+CTH GC V+K V+R + D V+TTY
Sbjct: 220 QTRSQVDILDDGYRWRKYGQKAVKNNKFPRSYYRCTHQGCNVKKQVQRLTKDEGVVVTTY 399
Query: 455 EGKHNHDVPAARGSGNHSVTR 475
EG H H + + H +++
Sbjct: 400 EGVHTHPIEKTTDNFEHILSQ 462
Score = 75.5 bits (184), Expect = 6e-14
Identities = 42/116 (36%), Positives = 63/116 (54%), Gaps = 1/116 (0%)
Frame = +1
Query: 179 SMMMKTENSSSMQSFSPEIASVQTNHNNGFQSDYNNYQPQQQQLQPQTLNRRSDDGYNWR 238
+M+M + S + S E+ N NN + + +P+ Q ++ DDGY WR
Sbjct: 94 NMLMSQISGGSNTNVSDELGG-SGNSNNKKKGEKKVKKPRYA-FQTRSQVDILDDGYRWR 267
Query: 239 KYGQKQVKGSENPRSYYKCTYPNCPTKKKVER-SLDGQITEIVYKGTHNHPKPQAT 293
KYGQK VK ++ PRSYY+CT+ C KK+V+R + D + Y+G H HP + T
Sbjct: 268 KYGQKAVKNNKFPRSYYRCTHQGCNVKKQVQRLTKDEGVVVTTYEGVHTHPIEKTT 435
>TC233582 similar to UP|WR32_ARATH (P59583) Probable WRKY transcription
factor 32 (WRKY DNA-binding protein 32), partial (27%)
Length = 759
Score = 110 bits (274), Expect = 2e-24
Identities = 50/104 (48%), Positives = 70/104 (67%)
Frame = +1
Query: 363 DEPDSKRWKVEGENEGISAPGSRTVREPRVVVQTTSDIDILDDGYRWRKYGQKVVKGNPN 422
++P+ KR + + PG +T + VV T D+ I DGYRWRKYGQK+VKGNP+
Sbjct: 43 NDPEPKRRLNNSDLDTAVKPGKKT----KFVVHATKDVGISGDGYRWRKYGQKLVKGNPH 210
Query: 423 PRSYYKCTHPGCPVRKHVERASHDLRAVITTYEGKHNHDVPAAR 466
R+YY+CT GCPVRKH+E A + +A+I TY+G H+HD+P +
Sbjct: 211 FRNYYRCTTAGCPVRKHIETAVDNSKALIITYKGMHDHDMPVPK 342
Score = 79.7 bits (195), Expect = 3e-15
Identities = 45/137 (32%), Positives = 72/137 (51%), Gaps = 7/137 (5%)
Frame = +1
Query: 231 SDDGYNWRKYGQKQVKGSENPRSYYKCTYPNCPTKKKVERSLDGQITEIV-YKGTHNHPK 289
S DGY WRKYGQK VKG+ + R+YY+CT CP +K +E ++D I+ YKG H+H
Sbjct: 151 SGDGYRWRKYGQKLVKGNPHFRNYYRCTTAGCPVRKHIETAVDNSKALIITYKGMHDHDM 330
Query: 290 PQATRRNSSSSSSLPIPHSNHISNDIQ------DQSYATHGSGQMDSVATPENSSISIGD 343
P +R+ S+SL + +N++Q Q+ T D+ ++ +G
Sbjct: 331 PVPKKRHGPPSASLVAAAAPASTNNVQLKKTGLLQNQETEAQCLEDTEGELTGEALDLGG 510
Query: 344 DDFEQSSQKCRSGGDEL 360
+ +S++ S G E+
Sbjct: 511 EKAIESARTLLSIGFEI 561
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.309 0.125 0.366
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 28,574,222
Number of Sequences: 63676
Number of extensions: 483891
Number of successful extensions: 5580
Number of sequences better than 10.0: 282
Number of HSP's better than 10.0 without gapping: 4405
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 5211
length of query: 579
length of database: 12,639,632
effective HSP length: 102
effective length of query: 477
effective length of database: 6,144,680
effective search space: 2931012360
effective search space used: 2931012360
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 62 (28.5 bits)
Lotus: description of TM0058a.3


