

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0058a.1
(1183 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
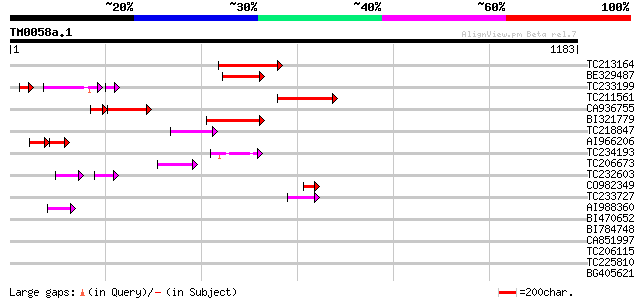
Score E
Sequences producing significant alignments: (bits) Value
TC213164 139 1e-32
BE329487 weakly similar to PIR|T14619|T146 reverse transcriptase... 127 2e-29
TC233199 91 7e-26
TC211561 weakly similar to UP|O22675 (O22675) Reverse transcript... 111 2e-24
CA936755 90 5e-24
BI321779 107 4e-23
TC218847 80 7e-15
AI966206 45 7e-12
TC234193 60 4e-09
TC206673 60 7e-09
TC232603 weakly similar to UP|Q39482 (Q39482) Legumin (Fragment)... 40 1e-08
CO982349 48 2e-05
TC233727 similar to UP|Q9FKH8 (Q9FKH8) Similarity to non-LTR ret... 48 2e-05
AI988360 44 4e-04
BI470652 similar to PIR|T17102|T171 RNA-directed DNA polymerase ... 43 0.001
BI784748 38 0.030
CA851997 37 0.067
TC206115 homologue to UP|CC48_SOYBN (P54774) Cell division cycle... 34 0.43
TC225810 homologue to UP|Q9ATF4 (Q9ATF4) Ribosomal protein L33, ... 32 1.7
BG405621 32 1.7
>TC213164
Length = 446
Score = 139 bits (349), Expect = 1e-32
Identities = 65/134 (48%), Positives = 94/134 (69%)
Frame = +3
Query: 436 LVGPFDEVEIKQAVWECGGTKSPGPDGFNFRFIQRFWDVLSGDVVKAVREFWMRGA*PRG 495
LV F++ E++ +W+ G KSPGPDG NF+FI+ FWD+L D+++ + EF+ G +
Sbjct: 18 LVARFEKKEVRATMWDRGNAKSPGPDGLNFKFIKEFWDILKTDLLRFLDEFYANGVFLKR 197
Query: 496 SNASFIVLIPKVKIPQLLNDFRPISLIGCIYKVVPKLFAIRLRLVIGKVIDERQFTFVGN 555
SNA F+ LIPKV P LN+++PISLIGCIYK+V KL + R + V+ +IDERQ F+
Sbjct: 198 SNAFFLALIPKVHDP*SLNEYKPISLIGCIYKIVAKLLSGRHKKVLPTIIDERQTVFMEG 377
Query: 556 RNMLDSVVVLNEVM 569
R+ L +VV+ NE+M
Sbjct: 378 RHTLHNVVIANEIM 419
>BE329487 weakly similar to PIR|T14619|T146 reverse transcriptase - beet
retrotransposon (fragment), partial (15%)
Length = 376
Score = 127 bits (320), Expect = 2e-29
Identities = 56/88 (63%), Positives = 68/88 (76%)
Frame = -2
Query: 445 IKQAVWECGGTKSPGPDGFNFRFIQRFWDVLSGDVVKAVREFWMRGA*PRGSNASFIVLI 504
IK AVW+CG KSPGPDG NF+FI+ FW+ L D+++ + EF G P+G NASFI LI
Sbjct: 375 IKSAVWQCGSVKSPGPDGLNFKFIKHFWERLKPDIIRFLNEFHANGIFPKGGNASFIALI 196
Query: 505 PKVKIPQLLNDFRPISLIGCIYKVVPKL 532
PKVK PQ LNDFRPISLIGC+YK+V K+
Sbjct: 195 PKVKHPQALNDFRPISLIGCVYKIVAKI 112
>TC233199
Length = 1105
Score = 91.3 bits (225), Expect(3) = 7e-26
Identities = 48/127 (37%), Positives = 69/127 (53%), Gaps = 5/127 (3%)
Frame = +2
Query: 71 LCIWNSKLFVLEESVDGPGFLGLMGRWGAAQQRCVIINIYSPCDIDGKRAMWDALCAWRG 130
LC + VLE+ V G GF+ L G W ++ I+NIYSPCD++ KR +W + R
Sbjct: 92 LCAYEVNHLVLEKKVIGSGFISLEGTWKGDGEKITIVNIYSPCDVNLKRNLWGQINQLRS 271
Query: 131 TCDVGEWCLAGDFNAVRYAEERRGSSGVLSSHRRE-----MREFNVFIANMDLVDIPLTG 185
WC+ DFN VR ER G+S +RE M EFN +I ++++ D+P G
Sbjct: 272 AS*NKLWCIVDDFNCVRRQSERVGAS------QREQGGGIMTEFNDWIVDLEVEDMPSVG 433
Query: 186 RRFTWRR 192
++TW R
Sbjct: 434 HKYTWYR 454
Score = 34.7 bits (78), Expect(3) = 7e-26
Identities = 13/30 (43%), Positives = 24/30 (79%)
Frame = +1
Query: 21 RELVCQQHVELLCIQETKCQVVDRRMCGLL 50
++LV ++ V++LC+QETK ++VD+ +C L
Sbjct: 7 KKLVVKEQVDMLCLQETKKEMVDKTVCQAL 96
Score = 31.2 bits (69), Expect(3) = 7e-26
Identities = 12/29 (41%), Positives = 16/29 (54%)
Frame = +1
Query: 201 IDRFLVSTSWCDVWPNCSHLVLNRDVSDH 229
+DR LVS W W +L+R+ SDH
Sbjct: 466 LDRVLVSVEWFSKWSGSIQFILDRNYSDH 552
>TC211561 weakly similar to UP|O22675 (O22675) Reverse transcriptase
(Fragment) , partial (59%)
Length = 1077
Score = 111 bits (277), Expect = 2e-24
Identities = 58/125 (46%), Positives = 77/125 (61%)
Frame = +1
Query: 559 LDSVVVLNEVMHEAKVKKRPSIFFKVDYEKAYNSVEWGFLEYMMGRMNFCAKWIGWIMGC 618
L S ++ NEV+ EAK + + FKVDYEKAY+SV W FL YMM RM+F KWI WI C
Sbjct: 1 LHSALIANEVIDEAKRSNKSCLVFKVDYEKAYDSVSWNFLMYMMWRMDFSPKWIKWIEEC 180
Query: 619 LNSATVSVLVNGSPSGEFHMEKGLRQGILLRRFFS*LLLKA*MGYSRMQLELGNLKVSRL 678
+ SA++SVLVNGSP+ EF ++GLRQG L F ++ + G R +E K +
Sbjct: 181 VKSASISVLVNGSPTAEFLPQRGLRQGDPLTPFLFNIVAEGLNGLMRRAVEENLYKPYLV 360
Query: 679 GQEAI 683
G +
Sbjct: 361 GANGV 375
>CA936755
Length = 424
Score = 90.1 bits (222), Expect(2) = 5e-24
Identities = 41/93 (44%), Positives = 56/93 (60%)
Frame = -2
Query: 204 FLVSTSWCDVWPNCSHLVLNRDVSDHCPLLMRQVVADWGPKPFRVLNCWLDDPRLFPFVE 263
FLVS W WP+ S L RD SDHCP++M+ DWGPKPFRV++ WL V
Sbjct: 294 FLVSEQWLLSWPDSSQYTLPRDFSDHCPIIMQTKKVDWGPKPFRVVDWWLHQKGYQRLVR 115
Query: 264 QSWKGMKIQGWGAYVLKEKLKGLRGSLRI*NKE 296
++W + GWG +LK KL+ L+ S++ +KE
Sbjct: 114 ETWSAEQQPGWGGILLKNKLRMLKLSIKQWSKE 16
Score = 40.8 bits (94), Expect(2) = 5e-24
Identities = 17/37 (45%), Positives = 25/37 (66%)
Frame = -1
Query: 168 EFNVFIANMDLVDIPLTGRRFTWRRANDQAQSRIDRF 204
EFN +I+ M+L +I G +TW R N + +SR+DRF
Sbjct: 403 EFNTWISEMELQEIKCVGSTYTWIRPNARVKSRLDRF 293
>BI321779
Length = 421
Score = 107 bits (266), Expect = 4e-23
Identities = 51/122 (41%), Positives = 78/122 (63%)
Frame = +2
Query: 410 FKSVAWLRPKLDGVQFSTITDRDNTELVGPFDEVEIKQAVWECGGTKSPGPDGFNFRFIQ 469
F+ + RPKL+GV+F + ++ DN L+ F+ EIK+ + +C +KSPGPDG++ I+
Sbjct: 5 FQEQIYGRPKLNGVEFLSFSNEDNAMLIQDFEVEEIKKVI*DCESSKSPGPDGYSLLLIK 184
Query: 470 RFWDVLSGDVVKAVREFWMRGA*PRGSNASFIVLIPKVKIPQLLNDFRPISLIGCIYKVV 529
FW+V+ +V+ +EF PRG+N SFI LI K P + + PISL+GC+YK+V
Sbjct: 185 IFWEVIKEEVINFFKEFHKFDFLPRGTNRSFIALIAKCDNPLDIGQYCPISLVGCLYKIV 364
Query: 530 PK 531
K
Sbjct: 365 *K 370
>TC218847
Length = 521
Score = 79.7 bits (195), Expect = 7e-15
Identities = 40/99 (40%), Positives = 59/99 (59%)
Frame = -3
Query: 335 LLIEFWRVLRYQESFLCQKACMKWLVEGDLNSGFFHSTINWKRRADSIVGLMVDGMWEDE 394
L+I W + ES L QKA KW+ E D NS +FH IN+ R +++ G+M+ G W +E
Sbjct: 297 LIIMEWTAAQAHESLLRQKARTKWIRERDCNSSYFHKLINYIHRTNAVNGVMIGGSWVEE 118
Query: 395 PVRVKAAIHDFFQKKFKSVAWLRPKLDGVQFSTITDRDN 433
P RVK A+ +FFQ +F + RP ++G F +I + N
Sbjct: 117 PNRVK-AVRNFFQIRFSESDYDRP*INGANFRSIGQQQN 4
>AI966206
Length = 502
Score = 45.4 bits (106), Expect(2) = 7e-12
Identities = 20/43 (46%), Positives = 27/43 (62%)
Frame = -2
Query: 83 ESVDGPGFLGLMGRWGAAQQRCVIINIYSPCDIDGKRAMWDAL 125
E V G GF+ L G W +R I+N Y+PCDI KRA+W+ +
Sbjct: 249 EEVVGSGFIMLEGIWKEDGERIAIVNTYAPCDIVLKRALWEEI 121
Score = 44.3 bits (103), Expect(2) = 7e-12
Identities = 18/41 (43%), Positives = 27/41 (64%)
Frame = -3
Query: 42 VDRRMCGLL*GDSEFEWKATPAVNRGGGLLCIWNSKLFVLE 82
+DR +C L GDSE W +PA+N GG+L IW+ + F ++
Sbjct: 371 IDRSICMELWGDSEVCWAESPAINTAGGVLSIWSEESFAMQ 249
Score = 32.7 bits (73), Expect = 0.97
Identities = 17/60 (28%), Positives = 28/60 (46%)
Frame = -1
Query: 12 GGNAKKRVVRELVCQQHVELLCIQETKCQVVDRRMCGLL*GDSEFEWKATPAVNRGGGLL 71
G K+ +R LV ++ V+++C+ ETK K +PA+N GG+L
Sbjct: 502 GDGVKRSSIRRLVLKEQVDMICLHETK--------------------KESPAINTAGGVL 383
>TC234193
Length = 697
Score = 60.5 bits (145), Expect = 4e-09
Identities = 40/113 (35%), Positives = 60/113 (52%), Gaps = 5/113 (4%)
Frame = -3
Query: 420 LDGVQFSTITDRDNTEL-----VGPFDEVEIKQAVWECGGTKSPGPDGFNFRFIQRFWDV 474
LDGV F TI DN L +G ++ W+ + +G+NF FI++ +V
Sbjct: 320 LDGVNFKTINGEDNDRLTHVLVLGTQNKRGSLGV*WK----QKFDSNGYNFNFIKKC*EV 153
Query: 475 LSGDVVKAVREFWMRGA*PRGSNASFIVLIPKVKIPQLLNDFRPISLIGCIYK 527
+ DVV+A++EF G RG+NASF+ P L DFRP++L+G + K
Sbjct: 152 MKEDVVRAIQEFHSHGCLSRGTNASFL-----THPP*GLGDFRPVTLVGSLCK 9
>TC206673
Length = 742
Score = 59.7 bits (143), Expect = 7e-09
Identities = 27/84 (32%), Positives = 47/84 (55%)
Frame = +3
Query: 308 VVQKINLLDVKEEEVGLQPEELVEMRDLLIEFWRVLRYQESFLCQKACMKWLVEGDLNSG 367
+ +K+N ++ L +E+ ++L + W ES L QK+ KWL EGD NS
Sbjct: 30 IKKKLNEVEDLASNRSLSVDEIKAKKELQQQLWEASTAYESLLRQKSRDKWLKEGDNNSA 209
Query: 368 FFHSTINWKRRADSIVGLMVDGMW 391
+FH+ IN++R + + G+++ G W
Sbjct: 210 YFHTVINFRRHYNGLQGILIQGEW 281
>TC232603 weakly similar to UP|Q39482 (Q39482) Legumin (Fragment), partial
(4%)
Length = 1123
Score = 40.4 bits (93), Expect(2) = 1e-08
Identities = 17/49 (34%), Positives = 29/49 (58%)
Frame = +2
Query: 178 LVDIPLTGRRFTWRRANDQAQSRIDRFLVSTSWCDVWPNCSHLVLNRDV 226
+V++ L GR++ W + + SR+DRFLV WPN + ++L R +
Sbjct: 524 VVNLALVGRKYNWYKDDKTTMSRLDRFLVYEE*LTTWPNMTLILLLRRI 670
Score = 38.1 bits (87), Expect(2) = 1e-08
Identities = 18/60 (30%), Positives = 26/60 (43%)
Frame = +1
Query: 95 GRWGAAQQRCVIINIYSPCDIDGKRAMWDALCAWRGTCDVGEWCLAGDFNAVRYAEERRG 154
G WG + + GK+ W+ + + WC+ GDFNAV EER+G
Sbjct: 304 GEWGRD*LKVFAVGFIVLVS*AGKKQEWEEIIRIKNEMGGERWCIVGDFNAVCSEEERKG 483
>CO982349
Length = 795
Score = 48.1 bits (113), Expect = 2e-05
Identities = 21/32 (65%), Positives = 26/32 (80%)
Frame = +3
Query: 614 WIMGCLNSATVSVLVNGSPSGEFHMEKGLRQG 645
WI GCL+SA+VS+LVN SP EF ++GLRQG
Sbjct: 69 WIRGCLHSASVSILVNESPINEFSPQRGLRQG 164
>TC233727 similar to UP|Q9FKH8 (Q9FKH8) Similarity to non-LTR retroelement
reverse transcriptase, partial (7%)
Length = 956
Score = 48.1 bits (113), Expect = 2e-05
Identities = 22/67 (32%), Positives = 40/67 (58%)
Frame = -3
Query: 579 SIFFKVDYEKAYNSVEWGFLEYMMGRMNFCAKWIGWIMGCLNSATVSVLVNGSPSGEFHM 638
++ K+D KA+++++ FL ++ + + + WI L SA +S+ VNG P G F
Sbjct: 768 NLALKIDIVKAFDTLDRDFLISVLNKFGYSHTFCNWIRVILLSAKLSISVNGEPVGFFSC 589
Query: 639 EKGLRQG 645
++G+RQG
Sbjct: 588 QRGVRQG 568
>AI988360
Length = 401
Score = 43.9 bits (102), Expect = 4e-04
Identities = 18/59 (30%), Positives = 30/59 (50%)
Frame = -3
Query: 79 FVLEESVDGPGFLGLMGRWGAAQQRCVIINIYSPCDIDGKRAMWDALCAWRGTCDVGEW 137
F L S G++GL G W Q + +N+YSPC+++ KR + + L + + W
Sbjct: 393 FSLNNSFQRDGYIGLEGTWKEGGQSVIFVNVYSPCELELKRRLSEELKSLKENSSCDHW 217
>BI470652 similar to PIR|T17102|T171 RNA-directed DNA polymerase (EC
2.7.7.49) (clone AmLi2) - garden snapdragon (fragment),
partial (22%)
Length = 111
Score = 42.7 bits (99), Expect = 0.001
Identities = 20/35 (57%), Positives = 26/35 (74%)
Frame = +1
Query: 618 CLNSATVSVLVNGSPSGEFHMEKGLRQGILLRRFF 652
CL+SA++S+LVNGSP+ EF +GLRQG FF
Sbjct: 7 CLSSASISILVNGSPTEEFKPXRGLRQGGPFGPFF 111
>BI784748
Length = 259
Score = 37.7 bits (86), Expect = 0.030
Identities = 17/35 (48%), Positives = 26/35 (73%)
Frame = +2
Query: 546 DERQFTFVGNRNMLDSVVVLNEVMHEAKVKKRPSI 580
D+RQ F+ R+ML S+++ NEV+ EAK ++PSI
Sbjct: 5 DKRQSAFIQGRHMLHSMMIANEVIDEAKRGQKPSI 109
>CA851997
Length = 636
Score = 36.6 bits (83), Expect = 0.067
Identities = 18/48 (37%), Positives = 25/48 (51%)
Frame = +1
Query: 460 PDGFNFRFIQRFWDVLSGDVVKAVREFWMRGA*PRGSNASFIVLIPKV 507
PDGFN F RFW + DV +A + GA P N + I +I ++
Sbjct: 316 PDGFNLGFYHRFWGMCGEDVFQACCMWLAEGAFPSSVNDTTIAIILEI 459
>TC206115 homologue to UP|CC48_SOYBN (P54774) Cell division cycle protein 48
homolog (Valosin containing protein homolog) (VCP),
partial (26%)
Length = 810
Score = 33.9 bits (76), Expect = 0.43
Identities = 9/29 (31%), Positives = 21/29 (72%)
Frame = +3
Query: 678 LGQEAILWCLYCSLLMILCFWVRLHFRIW 706
L + ++++C C+L+++ C+W+ L + IW
Sbjct: 690 LFEASLIFCFSCTLIILNCYWLNLKYHIW 776
>TC225810 homologue to UP|Q9ATF4 (Q9ATF4) Ribosomal protein L33, complete
Length = 736
Score = 32.0 bits (71), Expect = 1.7
Identities = 17/47 (36%), Positives = 21/47 (44%)
Frame = +1
Query: 594 EWGFLEYMMGRMNFCAKWIGWIMGCLNSATVSVLVNGSPSGEFHMEK 640
EW L MG+ + A W W C S +NGSPS H+ K
Sbjct: 322 EWKPLPLHMGQ-GYKASW*QWYCPCKIQVEPSTQINGSPSKSIHVSK 459
>BG405621
Length = 402
Score = 32.0 bits (71), Expect = 1.7
Identities = 14/26 (53%), Positives = 19/26 (72%)
Frame = +2
Query: 346 QESFLCQKACMKWLVEGDLNSGFFHS 371
+ES+ Q++ + WL GDLNS FFHS
Sbjct: 323 EESYWQQRSKIFWLKYGDLNSKFFHS 400
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.339 0.151 0.528
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 66,227,081
Number of Sequences: 63676
Number of extensions: 1168810
Number of successful extensions: 9964
Number of sequences better than 10.0: 50
Number of HSP's better than 10.0 without gapping: 9664
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 9948
length of query: 1183
length of database: 12,639,632
effective HSP length: 108
effective length of query: 1075
effective length of database: 5,762,624
effective search space: 6194820800
effective search space used: 6194820800
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.8 bits)
S2: 64 (29.3 bits)
Lotus: description of TM0058a.1


