

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0034a.1
(384 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
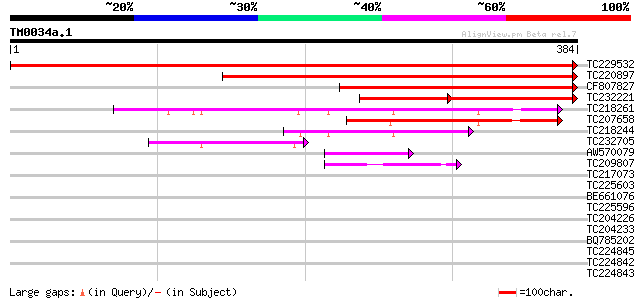
Score E
Sequences producing significant alignments: (bits) Value
TC229532 homologue to UP|GBA1_SOYBN (P49084) Guanine nucleotide-... 706 0.0
TC220897 homologue to UP|GBA2_SOYBN (P93163) Guanine nucleotide-... 465 e-131
CF807827 316 1e-86
TC232221 homologue to UP|GBA2_PEA (O04279) Guanine nucleotide-bi... 172 5e-74
TC218261 similar to UP|Q9LI02 (Q9LI02) ESTs C74776(E51022), part... 162 2e-40
TC207658 weakly similar to UP|Q9LI02 (Q9LI02) ESTs C74776(E51022... 113 2e-25
TC218244 similar to PIR|T51593|T51593 GTP-binding regulatory pro... 96 4e-20
TC232705 similar to UP|Q9LI02 (Q9LI02) ESTs C74776(E51022), part... 58 6e-09
AW570079 similar to PIR|S28875|S28 ADP-ribosylation factor 1 [im... 42 6e-04
TC209807 homologue to UP|Q9LFJ7 (Q9LFJ7) ADP-ribosylation factor... 41 8e-04
TC217073 homologue to UP|Q9LFJ7 (Q9LFJ7) ADP-ribosylation factor... 40 0.002
TC225603 homologue to UP|SARA_ARATH (O04834) GTP-binding protein... 39 0.003
BE661076 39 0.003
TC225596 homologue to UP|O81695 (O81695) Ras-like small monomeri... 39 0.003
TC204226 homologue to UP|SARA_ARATH (O04834) GTP-binding protein... 39 0.005
TC204233 homologue to UP|SARA_ARATH (O04834) GTP-binding protein... 39 0.005
BQ785202 38 0.007
TC224845 homologue to UP|Q70XK1 (Q70XK1) ADP-ribosylation factor... 38 0.009
TC224842 homologue to UP|Q70XK1 (Q70XK1) ADP-ribosylation factor... 38 0.009
TC224843 homologue to UP|Q8RUR2 (Q8RUR2) ADP-ribosylation factor... 38 0.009
>TC229532 homologue to UP|GBA1_SOYBN (P49084) Guanine nucleotide-binding
protein alpha-1 subunit (GP-alpha-1), complete
Length = 1469
Score = 706 bits (1821), Expect = 0.0
Identities = 347/384 (90%), Positives = 368/384 (95%)
Frame = +2
Query: 1 MGLLCSKNRRYNDADTEENAQTAEIERRIELETKAEKHIQKLLLLGAGESGKSTIFKQIK 60
MGL+CS++RR+ +A EENAQ AEIERRIELETKAEKHIQKLLLLGAGESGKSTIFKQIK
Sbjct: 5 MGLVCSRSRRFREAHAEENAQDAEIERRIELETKAEKHIQKLLLLGAGESGKSTIFKQIK 184
Query: 61 LLFQTGFDEAELKSYQPVIHANVYQTIKLLHDGAKELAQNDVDFSKYVISDENKEIGEKL 120
LLFQTGFDEAELKSY PV+HANVYQTIK+LHDG+KELAQND D SKYVIS+EN++IGEKL
Sbjct: 185 LLFQTGFDEAELKSYIPVVHANVYQTIKVLHDGSKELAQNDFDSSKYVISNENQDIGEKL 364
Query: 121 SEIGGRLDYPCLTKELALEIENLWKDAAIQETYARGNELQVPDCTHYFMENLHRLSDANY 180
SEIGGRLDYP LTKELA EIE LW+DAAIQETYARGNELQVPDC HYFMENL RLSDANY
Sbjct: 365 SEIGGRLDYPRLTKELAQEIETLWEDAAIQETYARGNELQVPDCAHYFMENLERLSDANY 544
Query: 181 VPTKDDVLYARVRTTGVVEIQFSPVGENKKSGEVYRLFDVGGQRNERRKWIHLFEGVSAV 240
VPTK+DVLYARVRTTGVVEIQFSPVGENK+SGEVYRLFDVGGQRNERRKWIHLFEGV+AV
Sbjct: 545 VPTKEDVLYARVRTTGVVEIQFSPVGENKRSGEVYRLFDVGGQRNERRKWIHLFEGVTAV 724
Query: 241 IFCAAISEYDQTLFEDENKNRMMETKELFEWVLKQPCFEKTSFMLFLNKFDIFEKKILKV 300
IFCAAISEYDQTL+EDENKNRMMETKELFEWVL+QPCFEKTSFMLFLNKFDIFEKK+L V
Sbjct: 725 IFCAAISEYDQTLYEDENKNRMMETKELFEWVLRQPCFEKTSFMLFLNKFDIFEKKVLNV 904
Query: 301 PLNVCEWFKDYQPVSTGKQEIEHAYEFVKKKFEESYFQNTAPDRVDRVFKIYRTTALDQK 360
PLNVCEWFKDYQPVSTGKQEIEHAYEFVKKKFEE YFQ+TAPD VDRVFKIY+ TALDQK
Sbjct: 905 PLNVCEWFKDYQPVSTGKQEIEHAYEFVKKKFEELYFQSTAPDCVDRVFKIYQATALDQK 1084
Query: 361 VVKKTFKLVDETLRRRNLFEAGLL 384
+VKKTFKLVDETLRRRNLFEAGLL
Sbjct: 1085LVKKTFKLVDETLRRRNLFEAGLL 1156
>TC220897 homologue to UP|GBA2_SOYBN (P93163) Guanine nucleotide-binding
protein alpha-2 subunit (GP-alpha-2), partial (62%)
Length = 1123
Score = 465 bits (1197), Expect = e-131
Identities = 226/240 (94%), Positives = 233/240 (96%)
Frame = +2
Query: 145 KDAAIQETYARGNELQVPDCTHYFMENLHRLSDANYVPTKDDVLYARVRTTGVVEIQFSP 204
KD AIQETYARG+ELQ+PDCT YFMENL RLSD NYVPTK+DVLYARVRTTGVVEIQFSP
Sbjct: 2 KDPAIQETYARGSELQIPDCTDYFMENLQRLSDTNYVPTKEDVLYARVRTTGVVEIQFSP 181
Query: 205 VGENKKSGEVYRLFDVGGQRNERRKWIHLFEGVSAVIFCAAISEYDQTLFEDENKNRMME 264
VGENKKS EVYRLFDVGGQRNERRKWIHLFEGVSAVIFCAAISEYDQTLFEDEN+NRM E
Sbjct: 182 VGENKKSDEVYRLFDVGGQRNERRKWIHLFEGVSAVIFCAAISEYDQTLFEDENRNRMTE 361
Query: 265 TKELFEWVLKQPCFEKTSFMLFLNKFDIFEKKILKVPLNVCEWFKDYQPVSTGKQEIEHA 324
TKELFEW+LKQPCFEKTSFMLFLNKFDIFEKKILKVPLNVCEWFKDYQPVSTGKQEIEHA
Sbjct: 362 TKELFEWILKQPCFEKTSFMLFLNKFDIFEKKILKVPLNVCEWFKDYQPVSTGKQEIEHA 541
Query: 325 YEFVKKKFEESYFQNTAPDRVDRVFKIYRTTALDQKVVKKTFKLVDETLRRRNLFEAGLL 384
YEFVKKKFEESYFQ+TAPDRVDRVFKIYRTTALDQKVVKKTFKLVDETLRR+NL EAGLL
Sbjct: 542 YEFVKKKFEESYFQSTAPDRVDRVFKIYRTTALDQKVVKKTFKLVDETLRRKNLLEAGLL 721
>CF807827
Length = 642
Score = 316 bits (810), Expect = 1e-86
Identities = 153/161 (95%), Positives = 157/161 (97%)
Frame = +3
Query: 224 RNERRKWIHLFEGVSAVIFCAAISEYDQTLFEDENKNRMMETKELFEWVLKQPCFEKTSF 283
R+ RKWIHLFEGVSAVIFCAAISEYDQTLFEDEN+NRM ETKELFEW+LKQPCFEKTSF
Sbjct: 78 RSTLRKWIHLFEGVSAVIFCAAISEYDQTLFEDENRNRMTETKELFEWILKQPCFEKTSF 257
Query: 284 MLFLNKFDIFEKKILKVPLNVCEWFKDYQPVSTGKQEIEHAYEFVKKKFEESYFQNTAPD 343
MLFLNKFDIFEKKILKVPLNVCEWFKDYQPVSTGKQEIEHAYEFVKKKFEESYFQ+TAPD
Sbjct: 258 MLFLNKFDIFEKKILKVPLNVCEWFKDYQPVSTGKQEIEHAYEFVKKKFEESYFQSTAPD 437
Query: 344 RVDRVFKIYRTTALDQKVVKKTFKLVDETLRRRNLFEAGLL 384
RVDRVFKIYRTTALDQKVVK TFKLVDETLRRRNLFEAGLL
Sbjct: 438 RVDRVFKIYRTTALDQKVVKNTFKLVDETLRRRNLFEAGLL 560
>TC232221 homologue to UP|GBA2_PEA (O04279) Guanine nucleotide-binding
protein alpha-2 subunit (GP-alpha-2), partial (38%)
Length = 778
Score = 172 bits (436), Expect(2) = 5e-74
Identities = 82/88 (93%), Positives = 87/88 (98%)
Frame = +2
Query: 297 ILKVPLNVCEWFKDYQPVSTGKQEIEHAYEFVKKKFEESYFQNTAPDRVDRVFKIYRTTA 356
+++VPLNVCEWFKDYQPVSTGKQEIEHAYEFVKKKFEE YFQ+TAPDRVDRVFKIYRTTA
Sbjct: 257 VVQVPLNVCEWFKDYQPVSTGKQEIEHAYEFVKKKFEELYFQSTAPDRVDRVFKIYRTTA 436
Query: 357 LDQKVVKKTFKLVDETLRRRNLFEAGLL 384
LDQK+VKKTFKLVDETLRRRNLFEAGLL
Sbjct: 437 LDQKLVKKTFKLVDETLRRRNLFEAGLL 520
Score = 124 bits (310), Expect(2) = 5e-74
Identities = 59/63 (93%), Positives = 61/63 (96%)
Frame = +1
Query: 238 SAVIFCAAISEYDQTLFEDENKNRMMETKELFEWVLKQPCFEKTSFMLFLNKFDIFEKKI 297
+AVIFCAAIS YDQTL+EDENKNRMMETKELFEWVLKQPCFEKTSFMLFLNKFDIFEKKI
Sbjct: 1 TAVIFCAAISGYDQTLYEDENKNRMMETKELFEWVLKQPCFEKTSFMLFLNKFDIFEKKI 180
Query: 298 LKV 300
L V
Sbjct: 181 LNV 189
>TC218261 similar to UP|Q9LI02 (Q9LI02) ESTs C74776(E51022), partial (35%)
Length = 1500
Score = 162 bits (410), Expect = 2e-40
Identities = 112/359 (31%), Positives = 178/359 (49%), Gaps = 55/359 (15%)
Frame = +3
Query: 71 ELKSYQPVIHANVYQTIKLLHDGAKELAQNDVDFSK------------------------ 106
EL+ + +I +N+Y+ + +L DG + + V
Sbjct: 3 ELQDVKLMIQSNMYKYLSILLDGRERFEEEAVSRMNGQGSPGQTMETGSNGEASNTSECI 182
Query: 107 YVISDENKEIGEKLSEI--GGRLD--YPCLTKELALEIENLWKDAAIQETYARGNELQ-V 161
Y ++ K + L +I G LD +P T+E A +E +W+D AIQET+ R +EL +
Sbjct: 183 YSLNPRLKHFSDWLLDIIATGDLDAFFPAATREYAPLVEEVWRDPAIQETFKRKDELHFL 362
Query: 162 PDCTHYFMENLHRLSDANYVPTKDDVLYARVRT--TGVVEIQFSPVGENKKSGEV----- 214
PD YF+ +S Y P++ D++YA T G+ ++FS KS
Sbjct: 363 PDVAEYFLSRAVEISSNEYEPSERDIVYAEGVTQGNGLAFMEFSLDDRIPKSDTYSENLD 542
Query: 215 --------YRLFDVGGQR-NERRKWIHLFEGVSAVIFCAAISEYDQTLFEDEN------- 258
Y+L V + NE KW+ +FE V AV+FC ++S+YDQ ++
Sbjct: 543 AQLPPLAKYQLIRVNAKGLNEGCKWVEMFEDVRAVVFCVSLSDYDQLSLSPDSSGSGTLV 722
Query: 259 KNRMMETKELFEWVLKQPCFEKTSFMLFLNKFDIFEKKILKVPLNVCEWFKDYQPVST-- 316
+N+M+++KELFE +++ PCF+ T +L LNK+DIFE+KI +V LN CEWF D+ PV
Sbjct: 723 QNKMVQSKELFETMVRHPCFKDTPLVLVLNKYDIFEEKISRVSLNTCEWFSDFCPVHAHH 902
Query: 317 GKQEIEH-AYEFVKKKFEESYFQNTAPDRVDRVFKIYRTTALDQKVVKKTFKLVDETLR 374
Q + H AY +V KF++ Y T + + + A D+ V + FK + E L+
Sbjct: 903 NNQSLAHQAYFYVAMKFKDLYASLTG-----KKLFVAQARARDRVTVDEAFKYIREVLK 1064
>TC207658 weakly similar to UP|Q9LI02 (Q9LI02) ESTs C74776(E51022), partial
(21%)
Length = 1028
Score = 113 bits (282), Expect = 2e-25
Identities = 58/151 (38%), Positives = 93/151 (61%), Gaps = 5/151 (3%)
Frame = +1
Query: 229 KWIHLFEGVSAVIFCAAISEYDQTLFED--ENKNRMMETKELFEWVLKQPCFEKTSFMLF 286
KW+ +FE V A+IFC ++S+YDQ + +N+++ +K+LFE ++K PCF+ T F+L
Sbjct: 154 KWLEMFEDVRAIIFCVSLSDYDQMWPTSTCQLRNKLLASKDLFESLVKHPCFKDTPFVLL 333
Query: 287 LNKFDIFEKKILKVPLNVCEWFKDYQPVST--GKQEIEH-AYEFVKKKFEESYFQNTAPD 343
LNK+D+FE KI KVPL+ CEWF D+ PV + H AY +V +F+E Y+ T
Sbjct: 334 LNKYDVFEDKINKVPLSTCEWFGDFCPVRPHHNNHALAHQAYYYVAMRFKELYYSLT--- 504
Query: 344 RVDRVFKIYRTTALDQKVVKKTFKLVDETLR 374
++ + +T D+ V + FK + E ++
Sbjct: 505 --NQKLFVGQTRGWDRSSVDEAFKYIREIIK 591
>TC218244 similar to PIR|T51593|T51593 GTP-binding regulatory protein
extra-large [validated] - Arabidopsis thaliana
{Arabidopsis thaliana;} , partial (19%)
Length = 1369
Score = 95.5 bits (236), Expect = 4e-20
Identities = 53/144 (36%), Positives = 85/144 (58%), Gaps = 15/144 (10%)
Frame = +1
Query: 186 DVLYARVRTT--GVVEIQFS-PVGENKKSGEV---------YRLFDVGGQR-NERRKWIH 232
D+LYA T+ GV ++FS P ++++ + Y+L V + E KW+
Sbjct: 283 DILYAEGVTSSNGVACVEFSFPQSVSEETVDTTDRYDSLVRYQLIRVHARGLGENCKWLE 462
Query: 233 LFEGVSAVIFCAAISEYDQTLFEDEN--KNRMMETKELFEWVLKQPCFEKTSFMLFLNKF 290
+FE V VIFC ++++YDQ + N+M+ +++ FE ++ P FE+ F+L LNKF
Sbjct: 463 MFEDVEMVIFCVSLTDYDQFSVDGNGCLTNKMVLSRKFFETIVTHPTFEQMEFLLILNKF 642
Query: 291 DIFEKKILKVPLNVCEWFKDYQPV 314
D+FE+KI +VPL CEWF D+ P+
Sbjct: 643 DLFEEKIEQVPLTKCEWFSDFHPI 714
>TC232705 similar to UP|Q9LI02 (Q9LI02) ESTs C74776(E51022), partial (12%)
Length = 390
Score = 58.2 bits (139), Expect = 6e-09
Identities = 35/115 (30%), Positives = 60/115 (51%), Gaps = 7/115 (6%)
Frame = +2
Query: 95 KELAQNDVDFSKYVISDENKEIGEKLSEIGGRLD----YPCLTKELALEIENLWKDAAIQ 150
+E A ++ S Y I+ K + L +I D +P T+E A ++ +W+D A+Q
Sbjct: 44 QETAADENKPSIYSINQRFKHFSDWLLDIMATGDLEAFFPAATREYAPMVDEIWRDPAVQ 223
Query: 151 ETYARGNEL-QVPDCTHYFMENLHRLSDANYVPTKDDVLYAR--VRTTGVVEIQF 202
ETY R EL +P+ YF++ +S Y P+ D+LYA ++ G+ ++F
Sbjct: 224 ETYKRRKELHNLPNVAKYFLDRAIEISSNEYEPSDKDILYAEGVTQSNGLAFMEF 388
>AW570079 similar to PIR|S28875|S28 ADP-ribosylation factor 1 [imported] -
Arabidopsis thaliana, partial (96%)
Length = 523
Score = 41.6 bits (96), Expect = 6e-04
Identities = 18/60 (30%), Positives = 33/60 (55%)
Frame = +2
Query: 214 VYRLFDVGGQRNERRKWIHLFEGVSAVIFCAAISEYDQTLFEDENKNRMMETKELFEWVL 273
++ ++DVGG R W H F+ +IF I++ D+ + ++ +RM+ EL + VL
Sbjct: 164 IFTVWDVGGHDKIRPLWRHYFQNTQGLIFVVDINDRDRVAYATDDLHRMLNEDELIDAVL 343
>TC209807 homologue to UP|Q9LFJ7 (Q9LFJ7) ADP-ribosylation factor-like
protein, complete
Length = 796
Score = 41.2 bits (95), Expect = 8e-04
Identities = 22/93 (23%), Positives = 44/93 (46%)
Frame = +1
Query: 214 VYRLFDVGGQRNERRKWIHLFEGVSAVIFCAAISEYDQTLFEDENKNRMMETKELFEWVL 273
++ ++DVGGQ R W H F +I+ + + ++ R+ + K+ F+ ++
Sbjct: 259 IFTVWDVGGQEKLRPLWRHYFNNTDGLIY----------VVDSLDRERIGKAKQEFQTII 408
Query: 274 KQPCFEKTSFMLFLNKFDIFEKKILKVPLNVCE 306
P + ++F NK D+ + P+ VCE
Sbjct: 409 NDPFMLNSVILVFANKQDL---RGAMTPMEVCE 498
>TC217073 homologue to UP|Q9LFJ7 (Q9LFJ7) ADP-ribosylation factor-like
protein, partial (98%)
Length = 1165
Score = 40.0 bits (92), Expect = 0.002
Identities = 23/93 (24%), Positives = 43/93 (45%)
Frame = +3
Query: 214 VYRLFDVGGQRNERRKWIHLFEGVSAVIFCAAISEYDQTLFEDENKNRMMETKELFEWVL 273
V+ ++DVGGQ R W H F +I+ + + ++ R+ + K+ F+ ++
Sbjct: 321 VFTVWDVGGQEKLRALWRHYFNNTDGLIY----------VVDSLDRERIGKAKQEFQTII 470
Query: 274 KQPCFEKTSFMLFLNKFDIFEKKILKVPLNVCE 306
P + ++F NK D+ K P +CE
Sbjct: 471 NDPFMLHSIILVFANKQDL---KGAMSPREICE 560
>TC225603 homologue to UP|SARA_ARATH (O04834) GTP-binding protein SAR1A,
complete
Length = 917
Score = 39.3 bits (90), Expect = 0.003
Identities = 24/90 (26%), Positives = 40/90 (43%), Gaps = 1/90 (1%)
Frame = +3
Query: 204 PVGENKKSGEV-YRLFDVGGQRNERRKWIHLFEGVSAVIFCAAISEYDQTLFEDENKNRM 262
P E G + ++ FD+GG + RR W + V AV++ L + +K R
Sbjct: 321 PTSEELSIGRIKFKAFDLGGHQIARRVWKDYYAKVDAVVY----------LVDSYDKERF 470
Query: 263 METKELFEWVLKQPCFEKTSFMLFLNKFDI 292
E+K+ + +L F++ NK DI
Sbjct: 471 AESKKELDALLSDESLTTVPFLILGNKIDI 560
>BE661076
Length = 595
Score = 39.3 bits (90), Expect = 0.003
Identities = 25/90 (27%), Positives = 41/90 (44%), Gaps = 1/90 (1%)
Frame = +2
Query: 204 PVGENKKSGEV-YRLFDVGGQRNERRKWIHLFEGVSAVIFCAAISEYDQTLFEDENKNRM 262
P E G++ ++ FD+GG + RR W + V AV++ + YD K R
Sbjct: 227 PTSEELSIGKIKFKAFDLGGHQIARRVWKDYYAQVDAVVY--LVDAYD--------KERF 376
Query: 263 METKELFEWVLKQPCFEKTSFMLFLNKFDI 292
E+K+ + +L F++ NK DI
Sbjct: 377 AESKKELDALLSDESLANVPFLVLGNKIDI 466
>TC225596 homologue to UP|O81695 (O81695) Ras-like small monomeric
GTP-binding protein, complete
Length = 1089
Score = 39.3 bits (90), Expect = 0.003
Identities = 25/90 (27%), Positives = 41/90 (44%), Gaps = 1/90 (1%)
Frame = +1
Query: 204 PVGENKKSGEV-YRLFDVGGQRNERRKWIHLFEGVSAVIFCAAISEYDQTLFEDENKNRM 262
P E G++ ++ FD+GG + RR W + V AV++ + YD K R
Sbjct: 289 PTSEELSIGKIKFKAFDLGGHQIARRVWKDYYAQVDAVVY--LVDAYD--------KERF 438
Query: 263 METKELFEWVLKQPCFEKTSFMLFLNKFDI 292
E+K+ + +L F++ NK DI
Sbjct: 439 AESKKELDALLSDESLASVPFLVLGNKIDI 528
>TC204226 homologue to UP|SARA_ARATH (O04834) GTP-binding protein SAR1A,
complete
Length = 1062
Score = 38.5 bits (88), Expect = 0.005
Identities = 24/90 (26%), Positives = 41/90 (44%), Gaps = 1/90 (1%)
Frame = +3
Query: 204 PVGENKKSGEV-YRLFDVGGQRNERRKWIHLFEGVSAVIFCAAISEYDQTLFEDENKNRM 262
P E G++ ++ FD+GG + RR W + V AV++ L + +K R
Sbjct: 393 PTSEELSIGKIKFKAFDLGGHQVARRVWKDYYAKVDAVVY----------LVDAFDKERF 542
Query: 263 METKELFEWVLKQPCFEKTSFMLFLNKFDI 292
E+K+ + +L F++ NK DI
Sbjct: 543 AESKKELDALLSDESLANVPFLVLGNKIDI 632
>TC204233 homologue to UP|SARA_ARATH (O04834) GTP-binding protein SAR1A,
complete
Length = 991
Score = 38.5 bits (88), Expect = 0.005
Identities = 24/90 (26%), Positives = 41/90 (44%), Gaps = 1/90 (1%)
Frame = +3
Query: 204 PVGENKKSGEV-YRLFDVGGQRNERRKWIHLFEGVSAVIFCAAISEYDQTLFEDENKNRM 262
P E G++ ++ FD+GG + RR W + V AV++ L + +K R
Sbjct: 300 PTSEELSIGKIKFKAFDLGGHQVARRVWKDYYAKVDAVVY----------LVDAFDKERF 449
Query: 263 METKELFEWVLKQPCFEKTSFMLFLNKFDI 292
E+K+ + +L F++ NK DI
Sbjct: 450 AESKKELDALLSDESLANVPFLVLGNKIDI 539
>BQ785202
Length = 423
Score = 38.1 bits (87), Expect = 0.007
Identities = 23/85 (27%), Positives = 39/85 (45%)
Frame = +2
Query: 208 NKKSGEVYRLFDVGGQRNERRKWIHLFEGVSAVIFCAAISEYDQTLFEDENKNRMMETKE 267
N + ++DVGGQ R W H F+ +IF ++ D+ + + +RM+ E
Sbjct: 95 NNNKNISFTVWDVGGQDKIRPLWRHYFQNTQGLIFVVDSNDRDRVVEARDELHRMLNEDE 274
Query: 268 LFEWVLKQPCFEKTSFMLFLNKFDI 292
L + VL ++F NK D+
Sbjct: 275 LRDAVL----------LVFANKQDL 319
>TC224845 homologue to UP|Q70XK1 (Q70XK1) ADP-ribosylation factor 1-like
protein, complete
Length = 865
Score = 37.7 bits (86), Expect = 0.009
Identities = 22/78 (28%), Positives = 38/78 (48%)
Frame = +2
Query: 215 YRLFDVGGQRNERRKWIHLFEGVSAVIFCAAISEYDQTLFEDENKNRMMETKELFEWVLK 274
+ ++DVGGQ R W H F+ +IF ++ D+ + + +RM+ EL + VL
Sbjct: 269 FTVWDVGGQDKIRPLWRHYFQNTQGLIFVVDSNDRDRVVEARDELHRMLNEDELRDAVL- 445
Query: 275 QPCFEKTSFMLFLNKFDI 292
++F NK D+
Sbjct: 446 ---------LVFANKQDL 472
>TC224842 homologue to UP|Q70XK1 (Q70XK1) ADP-ribosylation factor 1-like
protein, complete
Length = 854
Score = 37.7 bits (86), Expect = 0.009
Identities = 22/78 (28%), Positives = 38/78 (48%)
Frame = +1
Query: 215 YRLFDVGGQRNERRKWIHLFEGVSAVIFCAAISEYDQTLFEDENKNRMMETKELFEWVLK 274
+ ++DVGGQ R W H F+ +IF ++ D+ + + +RM+ EL + VL
Sbjct: 271 FTVWDVGGQDKIRPLWRHYFQNTQGLIFVVDSNDRDRVVEARDELHRMLNEDELRDAVL- 447
Query: 275 QPCFEKTSFMLFLNKFDI 292
++F NK D+
Sbjct: 448 ---------LVFANKQDL 474
>TC224843 homologue to UP|Q8RUR2 (Q8RUR2) ADP-ribosylation factor, partial
(48%)
Length = 1021
Score = 37.7 bits (86), Expect = 0.009
Identities = 22/78 (28%), Positives = 38/78 (48%)
Frame = +2
Query: 215 YRLFDVGGQRNERRKWIHLFEGVSAVIFCAAISEYDQTLFEDENKNRMMETKELFEWVLK 274
+ ++DVGGQ R W H F+ +IF ++ D+ + + +RM+ EL + VL
Sbjct: 290 FTVWDVGGQDKIRPLWRHYFQNTQGLIFVVDSNDRDRVVEARDELHRMLNEDELRDAVL- 466
Query: 275 QPCFEKTSFMLFLNKFDI 292
++F NK D+
Sbjct: 467 ---------LVFANKQDL 493
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.318 0.136 0.392
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,308,351
Number of Sequences: 63676
Number of extensions: 168574
Number of successful extensions: 873
Number of sequences better than 10.0: 68
Number of HSP's better than 10.0 without gapping: 864
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 867
length of query: 384
length of database: 12,639,632
effective HSP length: 99
effective length of query: 285
effective length of database: 6,335,708
effective search space: 1805676780
effective search space used: 1805676780
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0034a.1


